Abstract
The wild yak melanocortin-4 receptor (MC4R) gene was cloned and characterised in this study. Results showed that the wild yak MC4R gene was composed of 1343 bp nucleotides with no introns and a GC content of 49%, encoding a polypeptide with 332 amino acids. The speculated size of the open reading frame (ORF) was 999 bp with the start codon – ATG and stop codon – TAA. The sequence of the wild yak MC4R gene in this study is different from that reported before for the domestic yak, and represents a new haplotype within yak species. Nucleotide comparisons of the MC4R gene between wild yak and domestic yak, cattle and zebu showed relatively high sequence conservation. Two nucleotide mutations in the ORF region of the MC4R gene, namely C >T and C >A, existed in both yak species studied here. There were three nucleotide substitutions within the coding region that separated wild and domestic yak from cattle and zebu (A > G, C > G and C > G). One nucleotide mutation detected in the 3′ UTR separated wild yak from cattle and zebu (T > A) (no 3′ flanking region of domestic yak MC4R gene reported). There was no amino acid change between wild yak and domestic yak, or cattle and zebu. However, there were two changes between wild yak and domestic yak compared to cattle and zebu (A to T and V to L). A molecular phylogenetic tree showed wild yak and domestic yak to be joined first, followed by the cattle, zebu and then the sheep (Caprinae). The phylogenetic clustering result was not only identical to the taxonomy, but to the phylogenetic clustering using mitochondrial DNA.
Introduction
The wild yak (Bos grunniens mutus) is an endangered bovine found on the Qinghai-Tibetan Plateau at altitudes above 4500 m with number around 15,000 mostly living in China (Wiener et al. Citation2003). With a strong adaptability to the local ecological environment, wild yak can live freely and reproduce under severe conditions such as hypoxia, periods of short grass growth and a severely cold climate. Wild yak can hybridise with domestic yak (Bos grunniens) and cattle (Bos taurus) and improve their growth and adaptability (Wiener et al. Citation2003). The wild yak therefore possess an extremely precious gene pool.
The melanocortin-4 receptor (MC4R) gene has been identified as one of the candidate genes associated with an animal's meat quality, food intake and energy expenditure. As a result, the MC4R gene has been studied in 40 species at least. In a recent research, Cai et al. (Citation2011) sequenced the 999 bp open reading frame (ORF) of the domestic yak MC4R gene and defined two haplotypes based on the sequence nucleotide mutations. However, to date, no information on the molecular characterisation of the wild yak MC4R gene was available. Also, little is known of the difference between the wild yak MC4R gene and that of other species of the Bovinae subfamily. The objectives of this study were to (a) clone and characterise the wild yak MC4R gene, (b) analyse the genetic differences existing in MC4R at nucleotide and amino acid (AA) levels between wild yak and domestic yak, cattle and zebu and (c) explore the phylogenetic relationship among them based on the ORFs of MC4R gene.
Materials and methods
Genomic DNA extraction
This study used DNA from a wild yak living in Datong Breeding Farm of Qinghai province in China, which was captured in infancy on the Qinghai-Tibetan Plateau. Genomic DNA was extracted from the frozen blood sample using a standard phenol-chloroform method.
Primers and PCR amplification
A pair of primers was designed for PCR amplification of the wild yak MC4R gene. The primer sequences were PF 5′-GAA TCC AAA ATG AAC TCT ACC CA-3′ and PR 5′-CAA AGT GTC ACA TCC TTG TAA ATA A-3′. The PCR mixture contained 50–100 ng wild yak genomic DNA, 20 pM of each primer, 0.50 U ExTaq DNA polymerase (TakaRa, Dalian, China), 10×ExTaq Buffer (Mg2 + free), 0.25 mM dNTP, 2.5 mM MgCl2 and ddH2O in a final volume of 25 µL. The following cycles were applied: an initial denaturation at 95°C for 4 min, followed by 35 cycles at 95°C/1 min, 55.7°C/1 min and 72°C/1 min 30 s and a final extension at 72°C for 5 min.
Cloning and sequencing of wild yak MC4R gene
The PCR products of wild yak MC4R gene were electrophoresed and purified using a DNA Agarose Gel Extraction Kit (Omega). The purified fragments were cloned into pMD™18-T vector and subsequently transformed into Escherichia coli JM109 (TakaRa, Dalian, China). The recombinant plasmid DNA was obtained and then sequenced using an ABI 3730 automated sequencer (Applied Biosystems).
Sequence analysis and phylogenetic tree construction
The wild yak MC4R gene was characterised by referring to the MC4R gene information from other species available in GenBank. Nucleotide and deduced AA sequences of the MC4R gene between wild yak and other Bovinae species were aligned and analysed using BioEdit 7.0.9 (Hall Citation1999). Based on the ORFs of the MC4R gene, a Neighbour-joining (NJ) tree among Bovinae species was constructed using Mega 4.0 based on the Kimura's 2-parameter model with a bootstrap of 1000 replicates (Kumar et al. Citation2004).
Results and discussion
Structure features of the wild yak MC4R gene
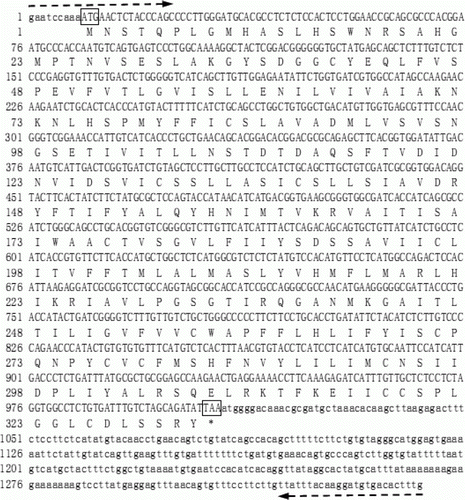
The complete sequence of the wild yak MC4R gene was deposited in GenBank (GenBank Accession No.: JF339983). The analysis results showed that the wild yak MC4R gene was composed of 1343 bp nucleotides with no introns and a GC content of 49%, encoding a polypeptide with 332 amino acids (). The speculated size of the ORF was 999 bp in length with the start codon – ATG and stop codon – TAA.
Figure 1. Sequences of nucleotide and deduced amino acid of wild yak MC4R gene. The open reading frame region and flanking region are shown by capital and lowercase, respectively. The positions of primers used for PCR are lined by broken horizontal arrows. The start codon ATG and stop codon TAA are boxed.
Nucleotide and amino acid variation
Nucleotide comparisons of the MC4R gene between wild yak and domestic yak, cattle and zebu showed a relatively high conservation rate. In the ORF region, the sequence similarities between wild yak and domestic yak, cattle and zebu were 99.8%, 99.6% and 99.5%, respectively. As the 3′ flanking region of domestic yak MC4R gene is not reported in Genbank, we here only compared the 3′ UTRs of wild yak with that of cattle and zebu, which had sequence similarities of 99.7% and 99.4%, respectively. Cai et al. (Citation2011) found two nucleotide mutations in the MC4R gene ORF region of the domestic yak population, namely C > T at position 60 and C > A at position 651; and defined two haplotypes. However, our research showed that the sequence of the wild yak MC4R gene sequenced here is different from that of the domestic yak reported by Cai et al. (Citation2011) (), indicating that this sequence represents a new haplotype within yak species. Moreover, three nucleotide substitutions within the coding region separated wild and domestic yak from cattle and zebu (A > G at position 31, C > G at position 856 and C > G at position 909). One nucleotide mutation detected in 3′ UTR separated wild yak from cattle and zebu (T > A at position 1118). The above results showed that although the MC4R gene has high similarities among Bovinae species, it still has enough genetic variation to differentiate within and between them.
Figure 2. Nucleotide sequence alignments of the MC4R ORFs and 3′UTRs among wild yak and other Bovinae species. ‘.’ and ‘-’ indicate that the nucleotide at that position compared with that of the wild yak are the same or a deletion, respectively. 3′UTRs are underlined. Differing nucleotide sites between wild yak, domestic yak and cattle, zebu are shaded grey, and among yak species are boxed. The start codon ATG and stop codon TAA are shaded black. Y represents C or T nucleotide, and R represents A or G nucleotide. Genbank Accession No.: wild yak (JF339983); domestic yak (a) (HM051375, HM051376 or HM051377); domestic yak (b) (HM051374); cattle (a) (EU366349); cattle (b) (EU366351, BC148892 or NM174110); zebu (EU366350).
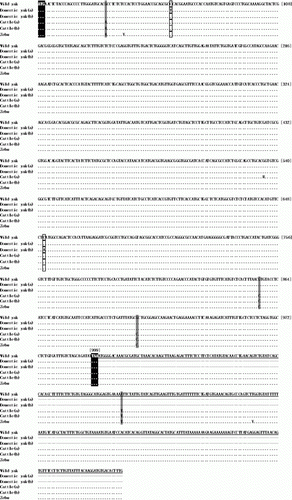
In our study, all four downloaded MC4R gene sequences for the domestic yak possessed the same deduced amino acid sequences, and the same was true for the cattle (). The similarities of deduced amino acid sequences between wild yak and domestic yak, cattle and zebu were 100%, 99.3% and 99.3%, respectively. There were no amino acid changes between wild yak and domestic yak, but two amino acid changes at position 11 and 286 differentiated wild yak and domestic yak from cattle and zebu (A to T and V to L). Although the wild yak are distinct from domestic yak in appearance [the wild yak is stronger, with a larger body, and covered in thick, long hair covering the whole body. It also has thick horns and slight differences in hair colour (Wiener et al. Citation2003)], our research indicates that mutations in the MC4R ORF are not a primary factor affecting their phenotypic differences. This implies that further research is necessary to compare the nucleotide variation in the flanking regions and gene regulation of the MC4R gene.
Phylogenetic relationship
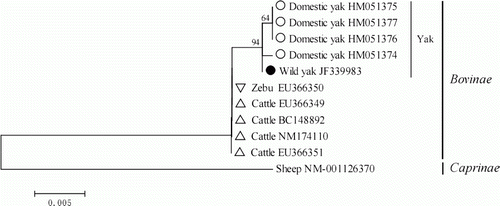
Phylogenetic research on the yak and other species of Bovinae is a key area in yak science (Li et al. Citation2006b). Based on the mtDNA D-loop region and Cyt b gene, previous research has explored the taxonomic relationship among wild yak, domestic yak and other species of Bovinae (Guo et al. Citation2006; Li et al. Citation2006a, 2007, 2008). In this study, a NJ tree was constructed using Mega 4.0 based on the ORF region of the MC4R gene of wild yak and corresponding sequences of domestic yak, cattle and zebu reported in GenBank. We used the sheep (O. aries) counterpart as an outgroup (GenBank accession number: NM_001126370) and the confidence of each branch was assessed with 1000 bootstrap replications (). The wild yak and domestic yak in the NJ tree were joined first, followed by the cattle, zebu and then the sheep. The result of phylogenetic clustering was not only identical to the taxonomy but also to the phylogenetic clustering using the mitochondrial DNA (Guo et al. Citation2006; Li et al. Citation2006a, Citation2007, Citation2008). The molecular phylogenetic tree constructed by the ORF regions of the MC4R gene can therefore reflect the evolutionary relationship of the bovines.
References
- Cai X , Mipam TD , Zhang HR , Yue BS . 2011 . Abundant variations of MC4R gene revealed by Phylogenies of Yak (Bos grunniens) and other mammals . Mol. Biol. Rep . 38 4 : 2733 – 2738 . doi: 10.1007/s11033-010-0418-2
- Guo SC , Liu JQ , Qi DL , Yang J , Zhao XQ . 2006 . Taxonomic placement and origin of yaks implications from analyses of mtDNA D-loop fragment sequences . Acta Theriol. Sin . 26 4 : 325 – 330 . [in Chinese, English abstract]
- Hall , TA . 1999 . BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT . Nucl. Acids Symp. Ser. , 41 : 95 – 98 .
- Kumar , S , Tamura , K and Nei , M . 2004 . MEGA: integrated software for molecular evolutionary genetics analysis and sequence alignment . Brief. Bioinform. , 5 : 150 – 163 .
- Li QF , Li YX , Zhao XB , Liu ZS , Zhang QB , Song DW , Qu XG , Li N , Xie Z . 2006a . Sequencing cytochrome b gene of mitochondrial DNA in yak and researching its origin and taxonomic status . Acta Vet. Zootech. Sin . 37 11 : 1118 – 1123 . [in Chinese, English abstract]
- Li QF , Zhao XB , Liu HL , Lin N , Xie Z . 2006b . A review of the research on taxonomic status in yak . Acta Zootax. Sin . 31 3 : 520 – 524 . [in Chinese, English abstract]
- Li , SP , Chang , H , Song , GM , Ma , GL , Chen , HY , Ji , DJ and Geng , RQ . 2007 . Molecular phylogeny and taxonomic status of domestic yak inferred from cytochrome B gene partial sequences . J. Anim. Vet. Adv. , 6 ( 12 ) : 1495 – 1499 .
- Li QF , Li YX , Zhao XB , Pan ZX , Liu ZS , Zhang QB , Qu XG , Song DW , Dong LY , Li N , et al. 2008 . Sequencing D-loop region of mitochondrial DNA in yak and study on its taxonomic status in Bovinae . Acta Vet. Zootech. Sin . 39 1 : 1 – 6 . [in Chinese, English abstract]
- Wiener , G , Han , JL and Long , RJ . 2003 . The yak , 2nd ed , Bangkok , , Thailand : Regional Office for Asia and the Pacific of the Food and Agriculture Organization of the United Nations .