Abstract
Paraflagellar rod (PFR), the major constituent proteins of flagellum, is restricted to kinetoplastids, euglenoids and dinoflagellates. Owing to their strategic location and invariable nature the proteins are considered as prospective vaccine targets. The present communication reports molecular cloning of the coding sequences of PFR1 and PFR2 genes, the two important constituent proteins of PFR of Trypanosoma evansi, Izatnagar isolate. The cloned nucleotide sequence of PFR1 revealed 99.8% sequence homology between the Izatnagar and China isolates with a single nucleotide change at position 867 bp of PFR1 open reading frame (ORF). The nucleotide sequencing data also revealed 99.8, 82.1, 79.9 and 72.9% sequence homology with Trypanosoma brucei, Trypanosoma cruzi, Leishmania infantum and Crithidia daenei, respectively. The cloned nucleotide sequence of PFR2 gene revealed 99.9% sequence homology between the Izatnagar and China isolates with a single nucleotide change at position 928 bp of PFR2 ORF of Izatnagar isolate. The PFR2 nucleotide sequence also showed 99.9, 82.4, 75.3 and 74.8% sequence homology with the published sequence of T. brucei, T. cruzi, L. infantum and C. fasciculata, respectively. The conserved nature of various PFR genes present in kinetoplastids could be exploited for development of a protective vaccine against multiple Trypanosoma species.
1. Introduction
Surra, caused by Trypanosoma evansi, is an economically important disease of a wide range of domestic and wild animals. The disease has most wide distribution and is a serious constraint for livestock production and health. The reports on the emergence of resistant strains to prevailing chemotherapeutic agents, potential toxic nature of the available drugs as well as environmental issues are some of the compelling reasons for development of a protective vaccine against the disease (Geerts et al. Citation2001; La Greca and Magez Citation2011). The quest for non-variable vaccine targets is emphasised because of the unique capacity of trypanosomes to undergo antigenic variation involving the surface glycoproteins and thereby defying the immunogenic benefits (Antoine-Moussiaux et al. Citation2009; Kurup and Tewari Citation2012). The unique structure of trypanosoma flagellum has made it an impressive vaccine target. The paraflagellar rod (PFR) extends alongside the axoneme from the flagellar pocket region up to the flagellum tip. The PFR is restricted to kinetoplastids, euglenoids and dinoflagellates and is vital for trypanosome motility (Bastin et al. Citation1998). However, in the amastigote form of T. cruzi and Leishmania spp. the reduced flagellum does not emerge from the flagellar pocket and, therefore, does not present PFRs (Portman and Gull Citation2010). The protein is unique among the kinetoplastids as their heteropolymers form the building block of flagellum (Abdille et al. Citation2008).The PFR is also known to provide support for metabolic regulators that may influence flagellar movement (Gadelha et al. Citation2005). Though there are more than 40 proteins associated with the structure of PFR, PFR1 and PFR2 are highly abundant (Portman and Gull Citation2010). The recombinant PFR2 from Leishmania mexicana (Saravia et al. Citation2004) and Trypanosoma cruzi (Luhrs et al. Citation2003) is shown to elicit strong immunogenicity in rodent models and pure PFR-like protein could protect mice against Trypanosoma cruzi infection (Ruth et al. Citation1995). The objective of the present study was to clone and sequence PFR proteins, PFR1 and PFR2 of T.evansi to investigate the degree of sequence homology existing between trypanosomes and other related organisms.
2. Materials and methods
2.1. Propagation and maintenance of parasites
The cryopreserved T. evansi (horse isolate) was propagated in vivo by intraperitoneal inoculation in experimental Swiss albino mice. At the height of parasitemia (108 trypanosomes/mL), the heart blood was collected from the exsanguinated mice using heparinised syringe. The host blood cell-free trypanosomes were harvested by DEAE-cellulose anion exchange chromatography (Lanham and Godfrey Citation1970).
2.2. RT-PCR amplification and cloning of PFR1 gene
Total RNA was extracted directly from the host cell-free trypanosomes, using Trizol reagent following the manufacturer's (Gibco BRL) recommendations. Complimentary DNA (cDNA) was synthesised from the total trypanosome RNA, using oligo dT primer following standard protocol. PCR-based amplification of the entire open reading frame (ORF) of PFR1 gene of T. evansi was done using self-designed specific primers (PFR1 forward primer (TEFPFR1F): 5′ ATG GCC GCA GTT GAC GAT G 3′ and PFR1 reverse primer (TEPFR1R): 5′ CTA TTC GAG GCG TGC CGG T 3′). The amplification cycle consisted of initial denaturation at 94°C for 4 min, followed by 32 cycles of denaturation at 94°C for 45 s, annealing at 57°C for 45 s and extension at 72°C for 2 min with a final extension at 72°C for 15 min. The amplified PCR product was visualised by ethidium bromide-stained agarose gel electrophoresis and isolated from the gel, using the QIA quick gel extraction kit (QIAGEN). The quantification of the purified PCR product was done spectrophotometrically (Nanodrop®, USA). A further confirmation of the PCR product was done by restriction enzyme analysis, using HindIII enzyme.
The purified PCR product was ligated in pGEM-T cloning vector, using standard protocol. The recombinant plasmid thus generated was designated as pGEM-T-PFR1 and was used for transformation of competent Escherichia coli DH5α cells. The transformed E.coli cells were cultured in LB agar media containing ampicillin at 37°C overnight. Positive clones were further selected in LB media containing ampicillin. Finally, the recombinant plasmid was extracted from the transformed DH5α cells, following standard protocol (Sambrook et al. Citation1989). For a confirmation of the presence of insert, a restriction digestion reaction was carried out using NcoI/Pst1 restriction enzymes, following standard protocol. The positive clone was custom sequenced for nucleotides.
2.3. Cloning of PFR2 gene
PFR2 gene was PCR amplified from the cDNA, using a specific pair of primers (Abdille et al. Citation2008) containing restriction sites for EcoRI and HindIII at the 5′ end of the forward (TEPFR2F) and reverse primer (TEPFR2R), respectively. The amplification cycle consisted of initial denaturation at 94°C for 4 min, followed by 32 cycles of denaturation at 94°C for 45 s, annealing at 57°C for 40 s and extension at 72°C for 2 min with a final extension at 72°C for 15 min. For the confirmation of specific amplification, the amplified product was electrophoresed in 1% ethidium bromide-stained agarose gel and subsequently ligated in pDRIVE T/A cloning vector, following standard protocol. The confirmation of the positive clones was done by colony PCR, using the same set of primers.
2.4. Nucleotide and amino acid sequence analysis of PFR1 and PFR2 genes
To find out the relative nucleotide sequence distances between the PFR1 gene of T. evansi and other PFR1 genes from kinetoplastids, a comparison of multiple nucleotide sequences was made using the ClustalW pair distance software package (DNASTAR, Madison, Wisconsin, USA). The coding sequence of PFR1 gene of T. evansi, Izatnagar isolate (GenBank Accession No. FJ968743) was compared with that of China isolate (GenBank Accession No.EU366960) of T. evansi as well as with that of Trypanosoma brucei (GenBank Accession No.XM838931), Trypanosoma cruzi (GenBank Accession No.AF005194), Leishmania. infantum (GenBank Accession.AY702344) and Crithidia daenei (GenBank Accession No.AY785777). Further, a comparison was made at the deduced amino acid sequence level.
Similarly, the nucleotide sequence of T. evansi, Izatnagar isolate PFR2 (GenBank Accession No.FJ901341) was compared with that of T. evansi, China isolate (GenBank Accession No.EU258755) as well as with T. brucei (GenBank Accession No.XM 842238), T. cruzi (GenBank Accession No.FJ222461), L. infantum (GenBank Accession No.XM001464593) and C. fasciculata (GenBank Accession No.AY568293). Further, a comparison was made at the deduced amino acid sequence level.
2.5. Isolation of genomic DNA
Genomic DNA of T. evansi was isolated from host cell-free trypanosomes by phenol: chloroform extraction method (Sambrook and Russell Citation2001).
2.6. Amplification of PFR1 and PFR2
In order to study the intron-less nature of the gene sequences, the target coding sequences for PFR1 and PFR2 were also amplified using the whole genomic DNA of T. evansi (horse isolate) as template using the specific forward and reverse primers.
3. Results and discussion
3.1. Cloning of PFR1 gene and its characterisation
The entire ORF of PFR1 gene was amplified from the Izatnagar isolate of T. evansi, using specific forward and reverse primers designed from the sequence information available in GenBank (Accession No: EU366960). The size of the single amplification product of 1770 bp was checked by agarose gel electrophoresis. The specificity of the purified amplified product was confirmed by restriction analysis using HindIII enzyme. The restriction digestion yielded two fragments of 1003 bp and 767 bp since a restriction site for HindIII is present at position 1003 bp (). The concentration of the purified PCR amplicon was measured as 35.2 ng/µl. This purified product of 1770 bp was ligated in a T/A cloning vector to facilitate its sequencing and characterisation.
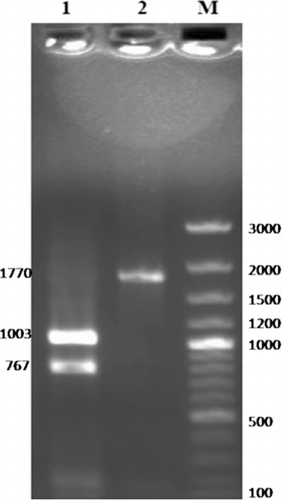
The insert was released from the plasmids extracted from the overnight grown cells of the positive clone by double restriction digestion, using NcoI and PstI enzymes. The positive clones were further checked for insert by colony PCR and custom sequenced for nucleotides. The nucleotide sequence revealed 99.9% homology with T. evansi, and China isolate with a single nucleotide substitution at position 867 bp of PFR1 ORF. The nucleotide sequence further showed sequence homology of 99.8, 82.1, 79.9 and 72.9% with T. brucei, T. cruzi, L. infantum and C. daenei, respectively. The mature PFR1 protein comprised of 590 amino acid residues. The deduced amino acid sequence of T. evansi PFR1 revealed 99.7% sequence homology between Izatnagar and China isolates. It also showed 99.8, 92.7, 84.7, 82.4% sequence homology with T.brucei, T. cruzi, L. infantum and C. daenei, respectively.
The computer-simulated recombinant PFR1 three-dimensional models generated showed a single domain. The protein conformation consisted of beta sheets with intermittent alpha chains. The template was monomer in nature.
3.2. Cloning of PFR2 gene and its characterisation
The entire ORF of PFR2 gene was amplified as a single band of 1800 bp from the Izatnagar isolate of T. evansi, using specific primers designed from the complete sequence information available in GenBank (Accession No: EU258755). The concentration of the purified PCR amplicon was measured as 35 ng/µl. The identity of the PCR product was further confirmed by restriction analysis using SspI enzyme. The restriction digestion yielded two bands of 1248 bp and 552 bp since the restriction site for SspI was located at position 552 bp (). This purified product of 1800 bp was ligated in a T/A cloning vector to facilitate its sequencing and characterisation. For this, pDRIVE T/A cloning vector, having an multiple cloning site incorporated into a LacZ a peptide coding region, was chosen for easy selection of recombinant clones.
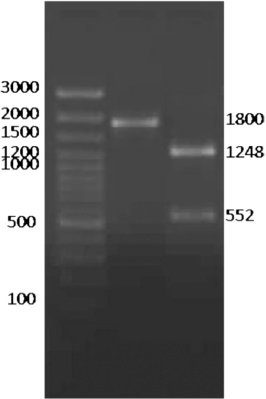
The insert was released by EcoRI restriction digestion of the plasmids extracted from the overnight grown cells of the positive clone. The release of the insert of desired length was checked by agarose gel electrophoresis of the enzyme-digested product. Colony PCR further confirmed the positive clone. The nucleotide sequencing revealed 99.9% homology between the Izatnagar and China isolates with a single nucleotide change at position 928 bp of PFR2 ORF. The nucleotide sequence also revealed a homology of 99.9, 82.4, 75.3 and 74.8% with T. brucei, T. cruzi, L. infantum and C. fasciculata, respectively. The mature PFR2 protein was comprised of 600 amino acid residues. The deduced amino acid sequence of T. evansi PFR2 revealed 99.8% sequence homology between Izatnagar and China isolates. It also showed 99.8, 90, 81.5, and 82% sequence homology with T. brucei, T.cruzi, L. infantum and C. fasciculata, respectively.
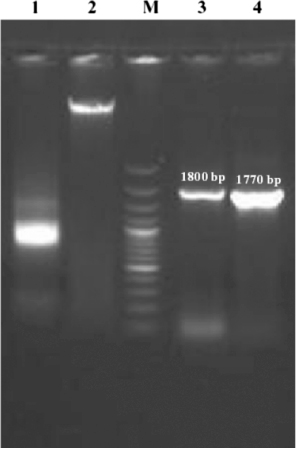
The sequence analysis of both PFR1 and PFR2 genes of T. evansi revealed the highly conserved nature of the gene in kinetoplastid species which coincides with the findings of Abdille et al. (Citation2008). Another significant finding was the absence of introns in the PFR1 and PFR2 coding sequences. The intron-less nature of the genes was further conformed by successful amplification of the target sequences coding for PFR1 and PFR2 from the whole genomic DNA template of T. evansi (horse isolate). Similar-sized amplicons for both PFR1 and PFR2 genes confirmed the intron-less nature of these genes (). The PFR proteins are important for motility and viability of the parasite. The immunogenic nature of PFR proteins was studied earlier using PFR2 alone (Saravia et al. Citation2004) or co-administration with PFR1 against T. cruzi infection in mice (Luhrs et al. Citation2003). Moreover, it is important to note that the PFR bears no significant sequence homology with any proteins of human and livestock origin (Clark et al. Citation2005). Though there is no report available on the immunoprophylactic potential of these target molecules against T. evansi, the present findings only further reaffirm the notion that vaccination with PFR molecule(s) could be effective not only against different strains of a trypanosome species but also against other species of the same genus. It is also likely that a PFR DNA or protein subunit vaccine with multiple epitopes of PFR proteins could be more immunogenic and effective against trypanosome infections than a monovalent PFR vaccine (Abdille et al. Citation2008). Therefore, the immunogenic and protective potential of PFR proteins of T.evansi need to be explored in laboratory and in large experimental animal models.
Acknowledgement
The authors acknowledge Indian Council of Agricultural Research, New Delhi, for awarding financial support in the form of Junior Research Fellowship to the first author. The authors also thankfully acknowledge the facilities provided by the Director, Indian Veterinary Research Institute, Izatnagar.
References
- Abdille MH, Li SY, Suo X, Mkoji G. 2008. Evidence for the existence of paraflagellar rod protein 2 gene in Trypanosoma evansi and its conservation among other kinetoplastid parasites. Exp Parasitol. 118:614–618. 10.1016/j.exppara.2007.11.011
- Antoine-Moussiaux N, Cornet A, Cornet F, Glineur S, Dermine M, Desmecht D. 2009. A non-cytosolic protein of Trypanosoma evansi induces CD45-dependent lymphocyte death. PLoS ONE. 4:1–12.
- Bastin P, Sherwin T, Gull K. 1998. Paraflagellar rod is vital for trypanosome motility. Nature 391:548. 10.1038/35300
- Clark K, April K, Gennadiy KS, Lal S, Stryker AG. 2005. Cloning and expression analysis of two novel paraflagellar rod domain genes in Trypanosoma cruzi. Parasitol Res. 96:312–320. 10.1007/s00436-005-1370-2
- Gadelha C, Wickstead B, De Souza W, Gull K, Cunha-e-Silva N. 2005. Cryptic paraflagellar rod in endosymbiont-containing kinetoplastid protozoa. Eukaryot Cell. 5:516–525. 10.1128/EC.4.3.516-525.2005
- Geerts S, Holmes PH, Diall O, Eisler MC. 2001. African bovine trypanosomiasis: the problem of drug resistance. Trends Parasitol. 17:25–28. 10.1016/S1471-4922(00)01827-4
- Kurup SP, Tewari AK. 2012. Induction of protective immune response in mice by a DNA vaccine encoding Trypanosoma evansi beta tubulin gene. Vet Parasitol. 187:9–16. 10.1016/j.vetpar.2012.01.009
- La Greca F, Magez S. 2011. Vaccination against trypanosomiasis Can it be done or is the trypanosome truly the ultimate immune destroyer and escape artist? Human Vaccines. 7:1225–1233. 10.4161/hv.7.11.18203
- Lanham SH, Godfrey DG. 1970. Isolation of salivarian trypanosomes from man and other animals using DEAE-cellulose. Exp Parasitol. 28:521–534. 10.1016/0014-4894(70)90120-7
- Luhrs KA, Fouts DL, Manning JE. 2003. Immunization with recombinant paraflagellar rod protein induces protective immunity against Trypanosoma cruzi infection. Vaccine. 21:3058–3069. 10.1016/S0264-410X(03)00108-7
- Portman N, Gull K. 2010. The paraflagellar rod of kinetoplastid parasites: From structure to components and function. Int J Parasitol. 40:135–148. 10.1016/j.ijpara.2009.10.005
- Ruth AW, Mark JM, Jose LS, Jerry EM. 1995. Pure paraflagellar rod protein protects mice against Trypanosoma cruzi infection. Infect Immun. 63:122–125.
- Sambrook J, Fritch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual . 2nd ed. New York: Cold Spring Harbor Laboratory Press.
- Sambrook J, Russel DW. 2001. (3rd ed). Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Saravia NG, Hazbon MH, Osorio Y, Valderrama L, Walker J, Santrich C, Cortazar T, LeBowitz JH, Travi BL. 2004. Protective immunogenicity of the paraflagellar rod protein 2 of Leishmania mexicana. Vaccine. 23:984–995. 10.1016/j.vaccine.2004.07.044
