Abstract
To analyze the associations of polymorphisms of prion protein gene (PRNP) with reproduction traits in Chinese Holstein bulls, significant statistical results are found in sperm quality traits between individuals with different 23-bp in-del, 12-bp in-del, and 24-bp in-del polymorphic genotypes. The individuals with 12-bp and 23-bp insertion polymorphisms had lower ejaculation volume, the number of straw frozen sperm once ejaculation and the number of straw per year than that of deletion polymorphisms at this two sites(p < 0.05). The individuals with 24-bp in-in genotype had lower number of straw per year than that of 24-bp in-del genotype (p < 0.050). These results showed the trend that resistant cattle PRNP genotypes possibly had negative effects on production traits in bulls. Therefore, this study provided possible molecular markers for bull breeding in resistance selection.
1. Introduction
Prion diseases are widely known as transmissible spongiform encephalopathies (TSE), which are a group of fatal neurodegenerative illnesses afflicting a variety of mammal species and humans, including scrapie in sheep and goat, bovine spongiform encephalopathy (BSE) in cattle, and others. The etiology of TSE has not been fully elucidated, but the central molecular event of conformational conversion of cell prion protein (PrPC) into scrapie-affected prion protein (PrPSc) was believed to be the infectious agent, which plays a central role in the pathogenesis of these diseases (Prusiner Citation1998; Aguzzi et al. Citation2008). Therefore, the prion protein gene (PRNP) has been studied as a candidate gene for resistance to prion diseases. It had been proved that PRNP polymorphisms had a strong correlation with the resistance to sheep scrapie, and the correlation has been well documented for sheep (Luhken et al. Citation2004; Goldmann Citation2008; Sweeney & Hanrahan Citation2008; Eiden et al. Citation2011). In cattle, and the BSE is characterized by the accumulation of an abnormal and protease resistant isoform of prion protein (PrP) in the central nervous system. The cattle PRNP has been mapped on chromosome 13q17, and its genomic structure has been already described (Sander et al. Citation2004; Kashkevich et al. Citation2007). Until now, many polymorphisms of PRNP had been identified in cattle. Previous reports have shown that several PRNP polymorphisms were associated with resistance to BSE in cattle (Sander et al. Citation2005; Haase et al. Citation2007; Kashkevich et al. Citation2007). In particular, a 23-bp in-del polymorphism within the PRNP promoter region and a 12-bp in-del polymorphism within intron 1 seemed to affect the binding sites for the transcriptional factors RP58 and SP1, respectively, and might influence PRNP expression (Sander et al. Citation2004; Sander et al. Citation2005; Xue et al. Citation2008). Furthermore, an octapeptide repeat or 24-bp in-del in the open-reading frame might be associated with resistance to BSE (Castilla et al. Citation2005; Brun et al. Citation2007). The research on transgenic mice expressing the bovine PrP suggested that the PrP with smaller numbers of octa/nona-peptide repeats increased BSE resistance (Brun et al. Citation2007). These results promoted widely investigation of the PRNP polymorphisms in many cattle breeds in the world and further evaluated the correlation between PRNP polymorphisms and the BSE resistance (Kim et al. Citation2009), and included several native Chinese breeds (Zhang et al. Citation2004; Zhao et al. Citation2010; Qin et al. Citation2011). On the other hand, the sheep's PRNP polymorphisms had effects on reproduction traits in some certain sheep breeds (Casellas et al. Citation2007; Lipsky et al. Citation2008). However, little information was available on the association between PRNP polymorphisms and the production performances in bulls. The present study was aimed to investigate the effects of PRNP polymorphisms on sperm quality in Chinese Holstein bulls, including 23-bp in-del in promoter region, 12-bp in-del in intron 1, and 24-bp in-del of octapeptide repeat in encoding region.
2. Materials and methods
2.1. Samples and information collection
A total of 98 sperm samples were collected from Shandong Province animal husbandry service center. Holstein bulls were 4–5 years old and fed with a mixture of roughage and concentrated provender. Hay and concentrated feed were added according to 1% and 0.5% of body weight, respectively. The sperm was collected twice per week by artificial pseudovagina except for August, which was the hottest month in this region.
The collected sperm was immediately stored at 37°C in a water bath for estimating the fresh sperm motility and concentration by light microscope (NIKON OPTIPHOT-2) and sperm concentration densitometer (Accucell 1145) in sperm inspection laboratory, the dilution volume, and the number of subpackaged frozen straw were auto estimated in this procedure. Then it was diluted and subpackaged in 0.25 mL, using frozen sperm straw distributor after 45 min for cold balancing at 4°C. The frozen sperm straw was stored in liquid nitrogen for next use after cryopreserved by GCS-300 programmed refrigerator (Japanese SHINKO Company). The frozen straw was taken out from liquid nitrogen after 24 h and was thawed in 38°C water bath for 10 sec, then the frozen sperm motility was estimated by light microscope monitor as mentioned above. For each bull, the protocol data of sperm quality has been available for two and more years.
2.2. DNA extraction and polymorphisms analysis
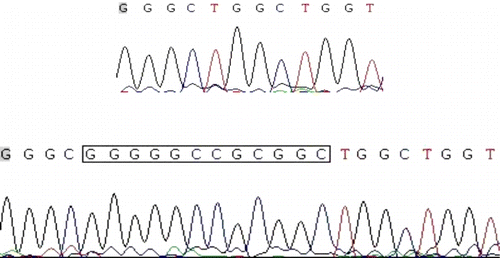
The collected sperm samples were delivered back to laboratory in a nitrogen canister. Genomic DNA was extracted using standard phenol-chloroform extraction protocol with minor modification. Subsequently, the concentration and the purity of each DNA sample was determined by NanoDrop 2000 (Thermo Fisher Scientific). Then, the DNA samples were genotyped for the 12-bp, 23-bp, and 24-bp polymorphisms by polymerase chain reaction (PCR), as reports described (Juling et al. Citation2006; Zhao et al. Citation2010). Moreover, the PCR products with different patterns were cloned into the pGEM-T vector for sequencing. Furthermore, the 261-bp and 285-bp PCR products at 24-bp in-del site mean the number of octapeptide repeat was 5 and 6, respectively (Zhao et al. Citation2010).
2.3. Statistical analysis
Allele frequencies were calculated by direct counting. The sperm quality traits data were presented as mean ± SD. The sperm motility was analyzed by one-way analysis of variance (ANOVA). The value of p < 0.05 was set as statistically significant.
3. Results
3.1. Genotyping and sequencing
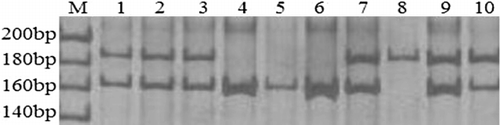
The PCR products were separated with electrophoresis, and the DNA fragment patterns in the gels could be genotyped clearly according to fragment length. The PRNP polymorphisms contained three, three, and two kinds of electrophoretograms at 12-bp, 23-bp and 24-bp in-del polymorphic sites, respectively. The results obtained by electrophoresis completely agreed with the sequencing results. The part of electrophoresis and sequencing results were shown in and .
3.2. PRNP polymorphisms at three sites
The 12-bp insertion and deletion allele frequencies were 39% and 71%, respectively. The 23-bp insertion and deletion frequencies were 51.6% and 48.4%, respectively. The 24-bp insertion and deletion frequencies were 96.3% and 3.7%, respectively. The genotype frequencies are shown in . The in-del heterozygous genotype had dominant frequencies at the 12-bp and 23-bp sites, which were 69.12% and 77.94%, respectively. The frequencies of these two heterozygous genotype were significantly higher than that of in-in and del-del homozygous genotypes. Furthermore, the frequency (92.65%) of in-in genotype was predominant at 24-bp polymorphic site.
Table 1. Comparison of semen motility with different PRNP genotypes in Holstein bulls.
3.3. Association of PRNP genotypes with sperm quality traits
The relationships between the polymorphisms of 12-bp, 23-bp, and 24-bp in-del and the sperm quality traits were analyzed, included fresh sperm motility, frozen sperm motility, ejaculation volume, the number of straw once ejaculation volume, and the number of straw per year. The results (in ) showed that these three polymorphisms had various effects on sperm quality traits including fresh sperm motility, ejaculation volume, the number of straw once ejaculation, and the number of straw per year. The bulls with 12-bp in-in genotype had lower ejaculation volume than that of the bulls with in-del and del-del genotypes (p < 0.05). The bulls with 23-bp in-in genotype had higher fresh sperm motility than that of the bulls with in-del genotypes (p < 0.05), but had the smallest number of straw once ejaculation and per year (p < 0.05). The bulls with 24-bp in-del genotype had the larger number of straw per year than that of the bulls with in-in genotypes (p < 0.05). No other significant difference was examined among sperm quality traits.
4. Discussion
PrP was detected from the BSE affected cattle for the first time in 1996 in the UK. Recently, it has made great progress on the studies of human and sheep PRNP polymorphisms and their correlation with humans and sheep resistance to prion diseases. It was reported that there were 60 polymorphic sites in the bovine PRNP that had been found before 2004 (Sander et al. Citation2004). And then, a total of 388 polymorphic sites had been localized, including 351 of the SNP types and 37 of the in-del types (Clawson et al. Citation2006). Among these PRNP polymorphic sites, a 23-bp in-del polymorphism in promoter and a 12-bp in-del polymorphism in intron 1 region had a relationship with resistance to BSE (Sander et al. Citation2005), a 24-bp in-del polymorphism in encoding region had a relationship not only with resistance to BSE, incubation period, but also with the prion diseases transmission rate (Derdowski et al. Citation2010). The analysis of sequence diversity in the PRNP of German cattle showed an association between 12-bp and 23-bp polymorphisms and resistance to BSE (Sander et al. Citation2004), especially for 12-bp in-del polymorphism which had main effects on resistance (Juling et al. Citation2006).
Previous reports revealed that the 12-bp insertion and 23-bp insertion polymorphisms all increased the resistance to BSE, while the 12-bp deletion polymorphism resulted in the increasing expression of PrP, and which resulted in higher susceptibility to prion diseases (Sander et al. Citation2005; Juling et al. Citation2006; Haase et al. Citation2007; Xue et al. Citation2008). The expression level of PrP had a negative relationship with the resistance to BSE, when the expression level of PrP was increasing, while the resistance would be decreasing. A functional analysis of the two in-del polymorphic sites revealed that the PrP expression level was lower when the two alleles were in-in genotype (Sander et al. Citation2005). Generally, the bovine with the low PrP expression level has an advantage on resistance to BSE, and increasing the frequencies of 12-bp insertion and 23-bp insertion alleles will be improving the resistance to BSE (Sander et al. Citation2005; Xue et al. Citation2008). Furthermore, it was reported that an increased number of octapeptide repeats in bovine PRNP would enhance host susceptibility to BSE, and the general number of repeats was five or six in previous reports (Castilla et al. Citation2005; Brun et al. Citation2007), and the number of it had four and/or seven in a few cattle (Sander et al. Citation2004; Seabury et al. Citation2004). These advances on PRNP polymorphisms provided theory guidance for cattle genetics and breeding. A national survey on Korean cattle was carried out based on the progress and the allelic, genotypic, and haplotypic distribution of PRNP was analyzed, and the resistance to BSE was further estimated (Kim et al. Citation2009). At present, selection for resistant PRNP genotypes and safety of animals are considered possible strategies for avoiding the prion diseases. On the other hand, the sheep resistant PRNP polymorphisms had different effects on reproduction performances in different breeds (Casellas et al. Citation2007; Lipsky et al. Citation2008), but the effects of PRNP polymorphisms on production performances in bulls had not been known, which are required to investigate.
In this preliminary study, our analysis revealed that the distributions of PRNP polymorphisms in Chinese Holstein bulls were according with the German cattle (including healthy and affected individuals) and similar with other reports in Holstein cattle, but different from other Chinese indigenous cattle breeds (Sander et al. Citation2004; Zhao et al. Citation2010; Qin et al. Citation2011). The results indicated that Chinese Holstein bulls had the middle resistance to BSE. The association analysis indicated that the 12-bp in-del polymorphism was associated with bull sperm ejaculation volume, and 23-bp in-del polymorphism was associated with fresh sperm motility, the number of straw per ejaculation and the number of straw per year. The polymorphisms of 12-bp in-in had an effect on resistance to BSE, but the individuals with this genotype had the lowest ejaculation volume. The cattle carried 23-bp in-in polymorphism had lower susceptibility, but the bulls had a smaller number of straws once ejaculation and the small number of straws per year. These results probably indicated that part of the resistant PRNP polymorphisms had a negative correlation with the bull sperm quality. The 24-bp in-del polymorphism had an effect on the number of octapeptide repeat in PrP, which affects the size of PrP complex and further affects transmission efficiency. The increasing number of repeat region would be shorten the BSE incubation period, vice versa (Brun et al. Citation2007; Derdowski et al. Citation2010). In current study, 5 of 98 cattle carried the 24-bp deletion were heterozygous genotypes and the number of octapeptide repeat was 6:5, which differed from previous reports in other native Chinese cattle breeds, but it was supposed that the octapeptide region may not play an important role in determining host resistance to BSE regardless of the number of repeat (Zhao et al. Citation2010). So we suggested that the three PRNP polymorphic sites could be potential genetic markers for sperm quality traits in bulls. However, the detailed associations between the PRNP polymorphisms and other economical traits remain elusive in bulls. Further investigation is necessary, in a large number of bulls and other cattle species. To date, occurrences of BSE have never been detected in China, but a breeding program for cattle with increasing resistance to BSE needs to consider the effects on reproduction and production performances in breeding. But unfortunately, our results showed that the resistant 12-bp and 23-bp in-in genotypes had negative effects on production traits in bulls, which can be overcome by raising more bulls.
Funding
This work was supported by the National Natural Science Foundation of China [No. 31001020]; the Scientific and Technological Innovation Team of Zhejiang Province [No. 2010R50027]; the Major Project of Science & Technology of Science Technology Department of Zhejiang Province, China [No. 2010C12007]; the National Natural Science Foundation of China [No. 31360540], the National High Technology Research and Development Program of China [863 Program, 2011AA100307].
Additional information
Funding
References
- Aguzzi A, Baumann F, Bremer J. 2008. The prion's elusive reason for being. Annu Rev Neurosci. 31:439–477. 10.1146/annurev.neuro.31.060407.125620
- Brun A, Gutiérrez-Adán A, Castilla J, Pintado B, Díaz-San Segundo F, Cano MJ, Alamillo E, Espinosa JC, Torres JM. 2007. Reduced susceptibility to bovine spongiform encephalopathy prions in transgenic mice expressing a bovine PrP with five octapeptide repeats. J Gen Virol. 88:1842–1849. 10.1099/vir.0.82568-0
- Casellas J, Caja G, Bach R, Francino O, Piedrafita J. 2007. Association analyses between the prion protein locus and reproductive and lamb weight traits in Ripollesa sheep. J Anim Sci. 85:592–597. 10.2527/jas.2006-308
- Castilla J, Gutiérrez-Adán A, Brun A, Pintado B, Salguero FJ, Parra B, Segundo FD, Ramírez MA, Rábano A, Cano MJ, Torres JM. 2005. Transgenic mice expressing bovine PrP with a four extra repeat octapeptide insert mutation show a spontaneous, non-transmissible, neurodegenerative disease and an expedited course of BSE infection. FEBS Lett. 579:6237–6246. 10.1016/j.febslet.2005.09.099
- Clawson ML, Heaton MP, Keele JW, Smith TP, Harhay GP, Laegreid WW. 2006. Prion gene haplotypes of U.S. cattle. BMC Genet. 7:51. 10.1186/1471-2156-7-51
- Derdowski A, Sindi SS, Klaips CL, DiSalvo S, Serio TR. 2010. A size threshold limits prion transmission and establishes phenotypic diversity. Science. 330:680–683. 10.1126/science.1197785
- Eiden M, Soto EO, Mettenleiter TC, Groschup MH. 2011. Effects of polymorphisms in ovine and caprine prion protein alleles on cell-free conversion. Vet Res. 4:30. 10.1186/1297-9716-42-30
- Goldmann W. 2008. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res. 39:30. 10.1051/vetres:2008010
- Haase B, Doherr MG, Seuberlic T, Drogemuller C, Dolf G, Nicken P, Schiebel K, Ziegler U, Groschup MH, Zurbriggen A, Leeb T. 2007. PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genet. 8:15. 10.1186/1471-2156-8-15
- Juling K, Schwarzenbacher H, Williams JL, Fries R. 2006. A major genetic component of BSE susceptibility. BMC Biol. 4:33. 10.1186/1741-7007-4-33
- Kashkevich K, Humeny A, Ziegler U, Groschup MH, Nicken P, Leeb T, Fischer C, Becker CM, Schiebel K. 2007. Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. FASEB J. 5:1547–1555. 10.1096/fj.06-7522com
- Kim Y, Kim JB, Sohn H, Lee C. 2009. A national survey on the allelic, genotypic, and haplotypic distribution of PRNP insertion and deletion polymorphisms in Korean cattle. J Genet. 88:99–103. 10.1007/s12041-009-0014-1
- Lipsky S, Brandt H, Lühken G, Erhardt G. 2008. Analysis of prion protein genotypes in relation to reproduction traits in local and cosmopolitan German sheep breeds. Anim Reprod Sci. 103:69–77. 10.1016/j.anireprosci.2006.12.005
- Luhken G, Buschmann A, Groschup MH, Erhardt G. 2004. Prion protein allele A136 H154Q171 is associated with high susceptibility to scrapie in purebred and crossbred German Merinoland sheep. Arch Virol. 149:1571–1580. 10.1007/s00705-004-0303-1
- Prusiner SB. 1998. Prions. P Natl Acad Sci USA. 95:13363–13383. 10.1073/pnas.95.23.13363
- Qin LH, Zhao YM, Bao YH, Bai WL, Chong J, Zhang GL, Zhang JB, Zhao ZH. 2011. Polymorphism of the prion protein gene (PRNP) in two Chinese indigenous cattle breeds. Mol Biol Rep. 38:4197–4204. 10.1007/s11033-010-0541-0
- Sander P, Hamann H, Drögemüller C, Kashkevich K, Schiebel K, Leeb T. 2005. Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 11:37408–37414. 10.1074/jbc.M506361200
- Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T. 2004. Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics. 5:19–25. 10.1007/s10048-003-0171-y
- Seabury CM, Honeycutt RL, Rooney AP, Halbert ND, Derr JN. 2004. Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. Proc Natl Acad Sci USA. 101:15142–15147. 10.1073/pnas.0406403101
- Sweeney T, Hanrahan JP. 2008. The evidence of associations between prion protein genotype and production, reproduction, and health traits in sheep. Vet Res. 39:28–46. 10.1051/vetres:2008004
- Xue G, Sakudo A, Kim CK, Onodera T. 2008. Coordinate regulation of bovine prion protein gene promoter activity by two Sp1 binding site polymorphisms. Biochem Biophys Res Co. 372:530–535. 10.1016/j.bbrc.2008.05.085
- Zhang L, Li N, Fan B, Fang M, Xu W. 2004. PRNP polymorphisms in Chinese ovine, caprine and bovine breeds. Anim Genet. 35:457–461. 10.1111/j.1365-2052.2004.01204.x
- Zhao H, Wang XY, Zou W, Zhang YP. 2010. Prion protein gene (PRNP) polymorphisms in native Chinese cattle. Genome. 53:138–145. 10.1139/G09-087