ABSTRACT
The solute carrier family 11 member A1 (SLC11A1), also known as natural resistance-associated macrophage protein 1 (NRAMP1), is a divalent transition metal (iron and manganese) transporter involved in iron metabolism and host resistance to certain pathogens. To study the genetic diversity of the SLC11A1 gene in Doom pigs of Assam especially on its evolution and differentiation within and among species, the partial sequence of the SLC11A1 gene was sequenced, characterized and compared with other published sequences of pigs and other species livestock. The gene sequence of Doom pigs showed the highest sequence identity with EF200584.1 (exotic pig) and the lowest similarity AY368475.1 (large white strain 008). One single nucleotide polymorphism (SNP) was identified in the heterozygous sequence at the 736 bp position (A→G). The sequence showed the highest sequence identity (81.74%) with that of O. aries and the lowest similarity (39.12%) with B. bubalis (Mehsana breed), respectively. Phylogenetic analysis revealed that the Doom pig is more closely related to EU135795.1 (Chinese local pig) and EF200584.1 (Pig, Iowa State University). The sequencing and phylogenetic analysis of the SLC11A1 gene of Doom pigs could be used for genetic selection with disease-resistance varieties and upgradation of indigenous germplasm of domestic livestock.
1. Introduction
Pig husbandry is of crucial importance in providing nutritional security and gainful employment to the weaker sections of society. Among the meat animals, pigs are the most prolific animals with shorter generation interval, high fecundity, high feed conversion efficiency and relatively require less space for housing. Doom variety of pigs is one of the indigenous pigs found in the lower part of Assam known for larger body size, high prolificacy and survival rate even in the low input system that is highly polymorphic in nature (Zaman et al. Citation2014). The solute carrier family 11 member A1 (SLC11A1) gene, formerly known as natural resistance-associated macrophage protein 1 (NRAMP1) encodes an integral membrane protein regulating the activity of macrophages (Blackwell Citation1996; Vidal et al. Citation1996; Forbes & Gros Citation2003). The gene also codes for an iron/divalent cation transporter (Blackwell et al. Citation2003) confer resistance to a number of antigenically different intracellular pathogens namely Salmonella typhimurium, Leishmania donovani and Mycobacterium bovis in mice (Vidal et al. Citation1993; Xiaoling et al. Citation2014). The SLC11A1 protein plays a pivotal role in the course of early infection by Mycobacteria, Leishmania and Salmonella (Cellier et al. Citation1995; Stear et al. Citation2001). SLC11A1 expression occurs mainly in macrophages and is interferon gamma-inducible. The report on phylogenetic analysis and sequencing of disease- resistance genes are very few in domestic animals. Keeping in view the above facts, the present study has been conducted to sequence and study the phylogenetic relationship of the SLC11A1 gene as a potent candidate gene for disease resistance in indigenous Doom pigs of Assam.
2. Material and methods
2.1. DNA extraction from blood
A total of 55 random blood samples of Doom pigs were collected from different districts of Assam, India, and genomic DNA were isolated by Rapid Isolation of Mammalian DNA method using standard protocol (Sambrook & Russell Citation2001).
2.2. Polymerase chain reaction (PCR)
A 934 bp region of the SLC11A1 gene was amplified by using a set of forward (F: 5′-TGAACACTTCATTTAACAGAAGA-3′) and reverse (R: 5′-GCCTCTGAGCAGGGAAGACT-3′) primers reported. PCR was performed in a thermocycler (Applied Biosystem, USA) in 25 μl reaction volume containing 1.0 µl DNA template, 0.5 µl each of forward and reverse primers of concentration 20 pmol/µl, PCR master mix 12.5 µl and nuclease-free water of 10.5 µl. PCR was carried out in 0.2 ml PCR tubes in a thermal cycler (Bio-Rad, Model-S100) under the following conditions: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 60 sec, extension at 72°C for 30 sec with a final extension step at 72°C for 5 min. The PCR products were separated by horizontal submarine agarose gel (2%) electrophoresis in 0.5x TAE buffer at 90 V and visualized using a gel documentation system.
2.3. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis
The PCR products (8.5 µl) were digested with 1.5 µl of Sma1 enzyme (Thermo-Scientific), 3.5 µl of 10x Tango buffer and 6.5 µl of nuclease-free water. The mixture was incubated at 30°C for 4 h and inactivation was done at 65°C for 20 min in a thermocycler (Bio-Rad, Model-S100). The digested products were analysed by 5% polyacrylamide gel electrophoresis (PAGE) and the gel was visualized in a GelDoc system (Bio-Rad). Identification of genotypes was done according to the restriction fragment patterns and it was confirmed by sequencing. WEB CUTTER 2.0 software was used for finding the restriction site of the SLC11A1 gene.
2.4. Sequencing and phylogenetic analysis
Sequencing of the PCR products (clustalW2.1) from different genotypes was done by using an automated DNA sequencer (SciGenom Private Ltd, Kakanad, Kochi, Kerala, India) and the nucleotide sequences obtained were confirmed through Basic Local Alignment search Tool analysis.
2.5. Nucleotide sequence comparison and phylogenetic analysis
The nucleotide sequences were aligned by using ClustalW2.1 software (http://www.ebi.ac.uk/Tools/clustalw/index.html), DNA Baser software was used for mutation analysis of the sequence and a phylogenetic tree was constructed from these alignments using the Neighbor-joining algorithm (clustalW2.1-phylogeny) with 1000 bootstrap replications. The sequences were submitted to National Center for Biotechnology Information. Five published sequences of the SLC11A1 gene of other breeds of pig, viz. two large white (AY368475 and AY368478), two Chinese local pig (EU135795 and AY556536) and one exotic pig (EF200584.1) were aligned and compared with the Doom pig sequence (KF 314185).
3. Results
3.1. Restriction site for the SLC11A1 gene
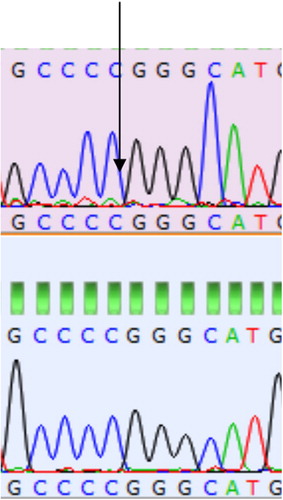
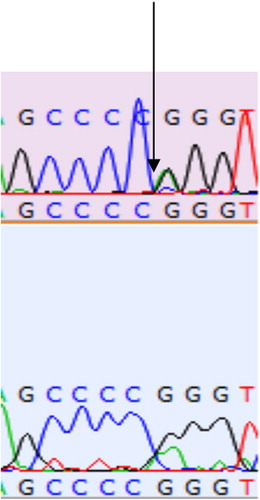
The restriction site for the SLC11A1 gene is found to be 505 and 736 bp. A 934 bp amplified product was obtained in respect of samples from all the Doom pigs from PCR amplification. At position 736 bp nucleotide transition (A ↔ G) was observed. Among the homozygous sequences single peaks () were observed with three fragments (505, 231 and 198 bp) and two fragments (505 and 429 bp), respectively, for both AA and BB genotypes while double peak () was observed among the sequences of all the heterozygous Doom pigs having four unique fragments (505, 429, 231 and 198 bp).
3.2. Percent identity at the nucleotide level with other pigs
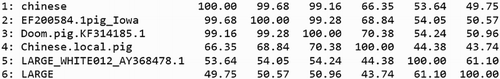
All the nucleotides of SLC11A1 genes obtained in the present study showed a 50–99.2% similarity with the other pigs. A similarity study of the SLC11A1 gene of Doom pigs showed that it has similarity of 99.28%, 99.16%, 70.38%, 54.24% and 50.96% with EF200584.1 (pig, Iowa State University), EU135795.1 (Chinese local pig), AY556536.1 (Chinese local pig), AY368478.1 (large white 012) and AY368475.1 (large white strain 008), respectively ().
3.3. Percent identity at the nucleotide level with other species
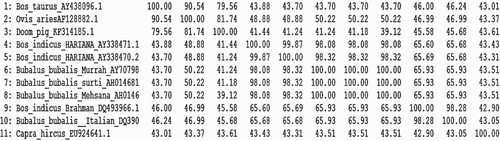
Sequence of Doom pigs is also compared with the published SLC11A1 gene sequences of other species namely cattle, buffalo (exotic and Indian) and sheep. A similarity study of the SLC11A1 gene of Doom pigs showed that it has similarity of 79.56%, 81.74%, 41.44%, 41.24%, 41.24%, 41.18%, 39.12%, 45.58%, 45.68% and 43.61% with AY438096.1 (Bos tauras), AF128882.1 (Ovis aries), AY338471.1 [Bos indicus (Hariana)], AY338470.2 [B. indicus (Hariana)], AY707989.1 [Bubalus bubalis (Murrah)], AH014681.1 [B. bubalis (Surti)], AH014682.1 [B. bubalis (Mehsana)], DQ493966.1 [B. indicus (Brahman)], DQ390205.1 [B. bubalis (Italian buffalo)] and Capra hircus, respectively ().
3.4. Phylogenetic analysis
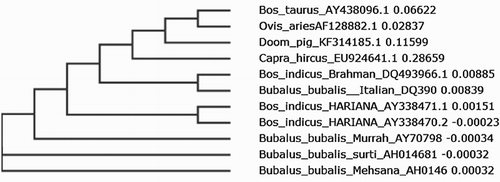
The nucleotide sequence of the SLC11A1 gene of Doom pigs was used for construction of a phylogenetic tree along with the published sequences of other pigs and different species of livestock. Study from the phylogenetic analysis revealed that the sequences. Doom pigs is somewhat more closely related to EU135795.1 (Chinese local pig) and EF200584.1 (exotic pig: ). Phylogenetic analysis with other species showed that Doom pigs are more closely related to B. tauras and O. aries and more distantly related to B. bubalis (Mehsana: ).
4. Discussion
Tuggle et al. (Citation2005) also reported similar findings, that is, A → G transition in Chinese indigenous pigs. However, they found in position 737 bp A → G transition compared to the present 736 bp position A ↔ G transition. Comparable findings were also reported by Liu et al. (Citation2011) who found A ↔ G transition at site 738 in intron1 forms another restriction site for the SmaI enzyme. Ding et al. (Citation2014) found one single nucleotide polymorphism (SNP) (g.130 C > T) in the exon1 and one SNP (g.657 A > G) in the intron1 region of the porcine NRAMP1 gene from DNA sequencing and PCR-RFLP analysis. Tuggle et al. (Citation1997) reported an NRAMP1 sequence with 538 amino acid protein and the derived pig protein sequence had much higher identity to NRAMP1 from humans, cattle and mice (87%, 88% and 85%, respectively).
Yakubu et al. Citation2014 had analysed the SLC11A1 gene to gain insight into the evolutionary proximity and divergence as well as the polymorphism of this gene in ruminants and non-ruminants including the attendant effects of the genetic variants on the function of the SLC11A1 protein. Thirty SLC11A1 gene sequences of six mammalian species classified as ruminant (goat, sheep, cattle, B. bubalis and Bubalus carabanensis) and non-ruminant (swine and horse) animals were investigated and it was found that the length of the SLC11A1 gene varied from 448 to 2357. They have also found that there was substantial genetic variation and polymorphism in the aligned sequences of the SLC11A1 gene within and across species. Functional analysis of non-synonymous mutations in cattle revealed that 25 of the amino acid substitutions at the peptide binding region E36G, T52A, N161S and V248I were likely to be beneficial while only Q312K was harmful.
5. Conclusion
The population of Doom pigs under study showed a diverged relationship with the other pig breeds and different species of animals in respect of the SLC11A1 gene. The sequence of Doom pigs showed high sequence identity with that of EF200584.1 and low similarity with AY368475.1 (large white strain 008). One SNP was also identified in the heterozygous sequence in the 736 bp position (A → G). The SLC11A1 gene sequence of Doom pigs showed high sequence identity (81.74%) with that of O. aries and low similarity with B. bubalis (Mehsana) (39.12%).
Acknowledgements
The authors are thankful to the Conservation Unit of Doom Pigs in Assam under the project ‘Conservation of threatened species’ for providing the samples used in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Blackwell JM. 1996. Structure and function of the natural resistance- associated macrophage protein (NRAMP1), a candidate protein for infectious and autoimmune disease susceptibility. Mol Med Today. 2:205–211.
- Blackwell JM, Searle S, Mohamed H, White JK. 2003. Divalent cation transport and susceptibility to infectious and autoimmune disease: continuation of the Ity/Lsh/Bcg/NRAMP1/SLC11A1 gene story. Immunol Lett. 85:197–203.
- Cellier M, Prive G, Belouchi A. 1995. NRAMP defines a family of membrane proteins. Proc Natl Acad Sci USA. 92:10089–10093.
- Ding X, Zhanga X, Yang Y, Ding Y, Xue W, Meng Y, Zhu W, Yin Z. 2014. Polymorphism, expression of natural resistance-associated macrophage protein 1 encoding gene (NRAMP1) and its association with immune traits in pigs. Asian-Australas J Anim Sci. 27(8):1189–1195.
- Forbes JR, Gros P. 2003. Iron, manganese, and cobalt transport by NRAMP1 (SLC11A1) and NRAMP2 (SLC11A2) expressed at the plasma membrane. Blood. 102:1884–1892.
- Liu Y, Qiu X, Xu J, Hu F, Li Y, Li H, Gong Y, Zhang Q. 2011. Association analysis between the polymorphism of the SLC11A1 gene and immune response traits in pigs. Asian J Anim Vet Adv. 6(6):580–586.
- Sambrook J, Russell D. 2001. Molecular cloning: a Laboratory manual. 3rd ed. Vol. 2. New York: Cold Spring Harbor Laboratory.
- Stear MJ, Bishop SC, Mallard BA, Raadsma H. 2001. The sustainability, feasibility and desirability of breeding livestock for disease resistance. Res Vet Sci. 71:1–7.
- Tuggle CK, Marklund L, Stabel TJ, Mellencamp MA, Stumbaugh A. 2005. Genetic markers for screening animals for improved disease resistance (NRAMP).United States Patent, 6844159B2. Available from: http://ddr.nal.usda.gov/handle/10113/6983
- Tuggle CK, Schmitz CB, Gingerich-Feil D. 1997. Rapid communication: cloning of a pig full-length natural resistance associated macrophage protein (NRAMP1) cDNA. J Anim Sci. 75(1):277.
- Vidal SM, Malo D, Vogan K, Skamene E, Gros P. 1993. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 73:469–485.
- Vidal SM, Pinner E, Lepage P, Gauthier S, Gras P. 1996. Natural resistance to intracellular infections: NRAMP1 encodes a membrane phospho-glycoprotein absent in macrophages from susceptible (NRAMP1 D169) mouse strains. J Immunol. 157(8):3559–3568.
- Xiaoling D, Xiaodong Z, Yong Y, Yueyun D, Weiwei X, Yun M, Weihua Z, Zongjun Y. 2014. Polymorphism, expression of natural resistance-associated macrophage protein 1 encoding gene (NRAMP1) and its association with immune traits in pigs. Asian-Australas J Anim Sci. 27:1189–1195.
- Yakubu A, Alade E, Dim NI. 2014. Molecular analysis of solute carrier family 11 member A1 (SLC11A1) gene in ruminants and non-ruminants using computational method. Genetika. 46(3):925–934.
- Zaman G, Laskar S, Ferdoci AM, Chandra Shekar M, Chetri AJ. 2014. Molecular characterization of Doom pigs using microsatellite markers. African J Biotechnol. 13(30):3017–3022.