?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Lipocalin-2 (LCN2) is a secreted protein, produced e.g. by neutrophils – the main group of cells involved in inflammatory conditions caused by pathogenic bacteria. So far, no studies have been carried out on the genetic variability of the LCN2 gene in cattle in terms of immunity/susceptibility to mastitis in cows. This study concerns the analysis of polymorphisms within the LCN2 gene in cattle and their impact on the production traits and the incidence of mastitis in Polish Holstein-Friesian cows. Using the PCR-RFLP method, two polymorphisms – g.98784892C > T and g.98784893T > G – were identified in intron 5 of the LCN2 gene, which represent two haplotypes (CT and TG, respectively). The CT haplotype was shown to have a significant impact on the average somatic cell score of milk, however, the CT and TG haplotypes were found to have no impact on the milk yield, protein, fat and lactose levels, or the incidence of mastitis in cows. An analysis of polymorphisms in the bovine LCN2 gene could be employed in future to support selection in dairy cattle breeding programmes focused towards the somatic cell count in milk, and the LCN2 gene itself could become a molecular marker of cows’ immunity to mastitis.
Introduction
Mastitis is a serious disease of dairy cattle. It increases the costs of milk production, causes substantial economic losses in most households worldwide, and reduces the technological quality of milk. It is also one of the most common reasons for the culling of heifers and cows (Aghamohammadi et al. Citation2018). An elevated somatic cell count in milk is usually a response of the immune system to inflammation of the mammary glands, which is mostly of bacterial origin, and as such it is considered an indicator of udder health status and an indicator of technological quality of milk (Sharma et al. Citation2011). Despite the rising standards of hygiene during milking, mastitis continues to be a global problem in the dairy cattle sector. As a result, researchers from all around the globe are seeking methods which could reduce the incidence of this disease. Susceptibility to mastitis can be a genetic trait, hence the search for genes which could be used as molecular markers of udder health status. The lipocalin-2 gene could be one of them (Pighetti and Elliott Citation2011; Abella et al. Citation2015).
Lipocalin-2 (LCN2, also known as siderocalin or neutrophil gelatinase-associated lipocalin, NGAL) belongs to a superfamily of small secreted proteins produced by various cells, including cells of the digestive, respiratory and immune systems, and cells of renal tubules. LCN2 was first discovered in granules of human neutrophils as a molecule associated with neutrophil gelatinase (Kjeldsen et al. Citation1993). It is a protein of molecular mass 25 kDa, composed of 198 amino acids (Flower Citation1996). LCN2 has an important role in immune response as it mediates various inflammatory processes and has pleiotropic effects. The serum level of this protein increases substantially at the time of an infection or inflammatory condition (Berger et al. Citation2005; Nairz et al. Citation2010). LCN2 helps to restrain the growth of microorganisms as a result of its capacity to bond to siderophores – compounds produced by bacteria, with a high specificity for chelating iron which is essential for bacterial growth (Schmidt-Ott et al. Citation2007). Not only does LCN2 protect against bacterial infections, but it also regulates the expression of pro-inflammatory cytokines in the body by reducing oxidative stress and maintaining iron homeostasis (Srinivasan et al. Citation2012). In addition to its bacteriostatic properties, lipocalin-2 has a variety of other important functions in the body, e.g. it regulates various cell processes such as cell apoptosis, cell migration/invasion, cell differentiation or tissue recovery. It seems that its role in the development of inflammatory processes is highly important as it was determined that secretion of LCN2 can be induced by IL-17 (interleukin 17), a cytokine produced by Th17 (T helper lymphocytes) cells, which participates in the pathogenesis of a number of autoimmune and inflammatory diseases (Shashidharamurthy et al. Citation2013).
Given the crucial role of lipocalin-2 in inflammatory processes, it was decided to analyse polymorphisms of the LCN2 gene in cattle and to investigate their association with immunity to mastitis in terms of the potential use as a mastitis marker. Another objective of the study was to determine the association between the analysed SNPs (single nucleotide polymorphisms) of the LCN2 gene and the composition of milk produced by Polish Holstein-Friesian cows.
Materials and methods
Samples collecting
The test material included milk samples collected into 100-mL sterile containers, which were subsequently used to isolate DNA (deoxyribonucleic acid) using the procedure developed by Pokorska et al. (Citation2016). The milk samples were collected from 298 cows (originating from 141 bulls) of the Polish Holstein-Friesian breed, black-and-white variety, in accordance with the ethical principles approved by the National Ethical Committee for Experiments on Animals at the Ministry of Science and Higher Education in Poland. The animals were kept in a free-stall system in a factory cattle farm in south-western Poland. The cows were fed a total mixed ration (TMR) and remained under constant veterinary supervision. Cases of clinical mastitis in cows was diagnosed by a veterinary doctor and information about them was gathered on the base of the records kept by the farm. During the diagnosis, the following aspects were taken into account: udder oedema, milk appearance, overall cow health, electrical conductivity of milk, individual somatic cell counts from quarters (SCC). The data on milk performance (daily milk yield, fat, protein and lactose levels, and somatic cell count, SCC) of the analysed cows were acquired based on monthly milk yield reports compiled in accordance with the recommendations of the International Committee for Animal Recording (ICAR).
PCR-RFLP procedure
The analysis included exon 5 of the bovine LCN2 gene and its flanking intron fragments (98784643–98784980 bp of the reference sequence, GenBank Accession No. NC_037338.1). The amplification of the 337-bp fragment of the LCN2 gene was performed using primers (F: 5′-TTCTGCAGACCCTCCATGA-3′, R: 5′-GTCTTCCTGACCCGGAGTG-3′) designed in Primer3plus (Untergasser et al. Citation2007). The PCR (polymerase chain reaction) was performed in a total volume of 50 µL, containing from 100 ng to 150 ng of genomic DNA, 1× PCR buffer with 200 µM of each dNTP (deoxynucleotide), 0.7 µM of each primer, 1% DMSO (dimethyl sulfoxide) and 0.04 U/µL of Phusion High-Fidelity DNA Polymerase (Thermo Scientific). The amplification was carried out under the following thermal conditions: initial denaturation at 98°C for 90 s, 35 cycles: at 98°C for 10 s, at 62°C for 20 s, at 72°C for 17 s, and the final elongation at 72°C for 6 min using C1000 Thermal Cycler (Bio-Rad).
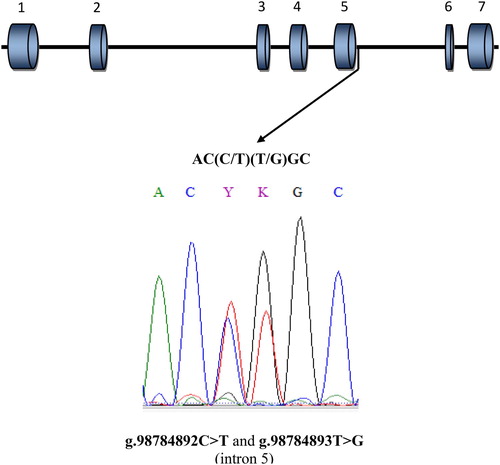
To identify SNPs in the analysed fragment of the LCN2 gene, amplicons were sequenced for 8 animals selected at random. All the sequencing was performed by a commercial company (Genomed S.A., Poland). Based on the sequenced samples, two polymorphisms – g.98784892C > T and g.98784893T > G (rs 208279129 and rs 135143347, respectively) were identified in the analysed fragment of intron 5 of the LCN2 gene (). To identify all the possible combinations of nucleotides at the identified polymorphic sites, the RFLP (restriction fragments length polymorphism) method was applied with the use of three endonucleases: Cfr10I, BveI and BseNI (Thermo Scientific, USA). The PCR products were digested separately by restriction enzymes in accordance with the manufacturer’s recommendations. The sizes of the obtainable RFLP fragments following the cleavage of the 337-bp fragment of the PCR product by restriction enzymes, for the SNPs identified in the analysed fragment of the LCN2 gene, are summarized in .
Table 1. The size of the RFLP fragments resulting from the cleavage of the 337-bp PCR product by restriction enzymes for the SNPs identified in the LCN2 gene.
The PCR-RFLP products were detected using 4% agarose gel (Sigma) stained with ethidium bromide (Sigma) in 1× TBE buffer and visualized under UV light using UVI-KS400I/Image PC (Syngen Biotech).
Statistical analysis
An analysis and a visualization of the haplotypes, and evaluation of the frequency of the resulting haplotypes formed by SNP variants of the LCN2 gene were performed using Haploview 4.2 (Barrett et al. Citation2005). The linkage disequilibrium (LD) between the pairs of alleles of the polymorphisms, given by the parameters D′ (standardized disequilibrium coefficient) and r2, was calculated using the same software.
The potential effect of the resulting haplotypes of the LCN2 gene on production traits of cows (averaged over the first three lactation periods), such as: average daily milk yield, average fat, protein and lactose levels in milk, and average somatic cell count in milk, was assessed using the PROC GLM procedure, SAS ® 9.4 software, with Scheffe’s test (SAS Institute, Cary NC, USA) according to the following model:
where Yij is the milk trait (average daily yield, average fat/protein/lactose, average somatic cell score (SCS)); µ is the overall mean; ai is the effect of the ith LCN2 haplotype combination (CT/CT, CT/TG and TG/TG), εij is the random error.
The GLM model (generalized linear model) included somatic cell score – a log-transformed somatic cell count (SCC) to normalize the distribution according to the following formula (Ali and Shook Citation1980):
The dominant and the additive effects of LCN2 haplotypes on the milk traits (average daily yield, average fat/protein/lactose, average SCS) were analysed using the PROC GLM procedure, SAS ® 9.4, using the following model:
where µ is the overall mean, b1 and b2 are additive and dominant regression coefficients, A1 is the additive effect, D2 is the dominant effect, εi is the random error.
To determine whether the identified haplotype is associated with the susceptibility or immunity of cows to mastitis, the incidence of mastitis in the cows was evaluated using a logistic regression model (PROC LOGISTIC, SAS Institute, version 9.4).
All the animals were divided into two groups: ‘immune to mastitis’ (class I) and ‘susceptible to mastitis’ (class II) according to the incidence of clinical mastitis. The group ‘immune to mastitis’ included animals which were found to have no mastitis or at most a single case of mastitis during the first three lactation periods. The group ‘susceptible to mastitis’ included cows affected by more frequent episodes of clinical mastitis (from 2 to 12 episodes recorded during the first three lactation periods).
The model of clinical mastitis was defined as follows:where P = p(Y = 1); Y is the occurrence of disease denoted as 1 – zero episodes or a single episode of clinical mastitis and 2 – at least two episodes of clinical mastitis during the first three lactation periods; b0 is the intercept; b1 is the regression coefficients of the disease occurrence on fixed effects; L is the combination of LCN2 haplotypes (CT/CT, CT/TG and TG/TG).
Results
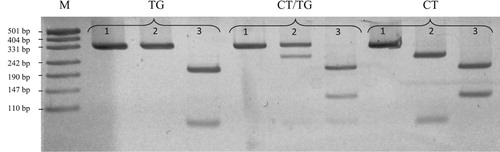
Based on the analysis of the sequenced samples, two polymorphisms – g.98784892C > T and g.98784893T > G – were identified in the analysed fragment of intron 5 of the LCN2 gene (). It was found that in each analysed loci there are two alleles and three genotypes. The most frequent genotype for g.98784892C > T polymorphic site was CC and for g.98784893T > G mutation was TT. In both case, the frequencies of these genotypes were amounted to 0.59. The frequencies of other genotypes were as follows: in case of g.98784892C > T: CT-0.37, TT-0.04, and in case of g.98784893T > G: TG-0.37, GG-0.04. The genotyping of the identified bi-allelic SNPs, with the use of three endonucleases, showed () that nucleotides at analysed sites always follow the same pattern (CT, CT/TG or TG) and that they form haplotype block Frequencies of these haplotypes were 0.775 for CT and 0.225 for TG.
Figure 2. The electrophoresis patterns of analysed LCN-2 haplotypes genotyping. In the first, second and third lines for each haplotype there is a band pattern resulting from cutting restriction endonuclease: Cfr10I, BveI and BseNI respectively, M – molecular marker pUC19DNA/MspI (Thermo Scientific).
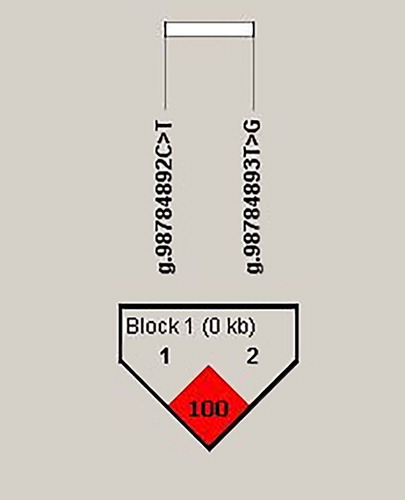
The analysis of the linkage disequilibrium (LD) demonstrated a 100% linkage between the analysed SNPs (D′ = 1, r2 = 1) (), and therefore only one of them needs to be genotyped to diagnose an allelic change.
Figure 3. Linkage disequilibrium (LD) plot in Holstein-Friesian cattle. Colour scheme is according to Haploview r2 scheme. Number in red cell stand for pairwise r2 -value (%) between the g.98784892C > T and g.98784893T > G SNPs.
The analysis of the resulting haplotypes of the LCN2 gene on production traits of cows showed that the CT/CT haplotype combination has a significant impact on the average somatic cell score of milk, which was significantly higher compared to the CT/TG haplotype combinations (). On the other hand, the haplotypes identified were found to have no significant impact, involving either additive or dominance effects, on the analysed milk traits ().
Table 2. Effect of LCN2 haplotype on milk of Holstein-Friesian cows.
Table 3. The additive and dominant effect of LCN2 haplotype on milk traits.
The analysis of logistic regression demonstrated no significant differences between the identified LCN2 haplotypes and the incidence of mastitis in the analysed herd of Holstein-Friesian cows (P value – 0.20).
Discussion
Lipocalin-2 is a protein which performs very important biological functions in the body, mostly in the extrinsic apoptotic pathway in the absence of a ligand (Devireddy et al. Citation2005), but it is also an active factor to the innate immune response, including in particular immune response to bacterial infections. This is due to its capacity to bond iron ions, thus absorption of this ions by bacteria is not possible and this mechanism prevents active growth of bacteria (Flo et al. Citation2004; Smith Citation2007). LCN2 interferes with the siderophore-mediated transport system by capturing bacterial siderophores produced by various, mostly pathogenic, bacteria such as Escherichia coli, which had a significant impact on susceptibility to infections and sepsis in mice with LCN2 deficiency (Xiao et al. Citation2017). Lipocalin-2 was also shown to exhibit anti-bacterial properties against other bacteria, e.g. Staphylococcus aureus (Robinson et al. Citation2014; Breyne et al. Citation2017).
Due to the fact, that mastitis in cows is mostly caused by bacteria such as Escherichia coli, Streptococcus agalactiae or Staphylococcus aureus (Holmberg et al. Citation2012; Jensen et al. Citation2013), it seems that lipocalin-2 should be an essential factor in combating such pathogens. Sharma et al. (Citation2016) showed that plasmid-mediated hLCN2 gene transfer into the bovine mammary epithelial stem cells can provide a source of hLCN2 and help to better protect the mammary glands against major mastitis-causing bacteria. Given that both production traits and susceptibility to mastitis are determined by enviromental and also genetic factors, analysis of polymorphisms of the LCN2 gene is reasonable. Our studies have shown that the haplotype identified in the analysed fragment of intron 5 of the LCN2 gene had a significant impact on the average somatic cell score of cow’s milk. The somatic cell count in milk is considered one of the fundamental indicators of udder health status in cows. The somatic cell count in milk obtained from healthy cows is less than 400,000/mL (Regulation EC Citation853/Citation2004). Schepers et al. (Citation1997) consider that the somatic cell count provides a threshold level between healthy and non-healthy udder. An increased somatic cell count (SCC) in cow’s milk is a response to the invasion of pathogens and the chronic inflammatory condition in the udder, and therefore SCC (or normalized somatic cell count – somatic cell score, SCS) is used for diagnosing mastitis.
Even though the SNPs studied by us are located in the non-coding fragment of the LCN2 gene, it does not mean that they have no impact on the gene functions. Polymorphisms located in introns can interfere with the process of mRNA (messenger ribonucleic acid) splicing, which in turn may lead to the formation of improper transcripts. Non-coding sequences may also include loci that are recognized by regulatory factors which, when bound, may affect the transcript level. Additionally, the most recent studies show that SNPs present in introns of one gene may affect the expression of other genes located at a considerable distance from them (million bp) (Patrushev and Kovalenko Citation2014).
Conclusions
In conclusion, based on an analysis of polymorphism in intron 5 of the bovine lipocalin-2 gene, it was shown that there are two CT and TG haplotypes. The haplotype was shown to have a significant impact on the average somatic cell score of cow’s milk, even though it was not associated with the incidence of mastitis in cows. Polymorphisms of the LCN2 gene in cattle should be investigated further as in future, this gene could become a good molecular marker of udder health status and help in developing a strategy for prevention of mastitis in dairy cattle herds, while improving milk performance.
Ethics approval
All animal procedures were approved by the National Ethical Committee for Experiments on Animals at Ministry of Science and Higher Education in Poland. The experiments were performed according to the regulations and guidelines established by this Committee.
Acknowledgements
The authors thank all staff at Faculty of Animal Sciences University of Agriculture in Krakow, who helped collect data for research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
ORCID
Joanna Pokorska http://orcid.org/0000-0003-3768-2837
Dominika Kułaj http://orcid.org/0000-0003-0164-2446
Additional information
Funding
References
- Abella V, Scotece M, Conde J, Gómez R, Lois A, Pino J, Gómez-Reino JJ, Lago F, Mobasheri A, Gualillo O. 2015. The potential of lipocalin-2/NGAL as biomarker for inflammatory and metabolic diseases. Biomarkers. 20:565–571. doi: 10.3109/1354750X.2015.1123354
- Aghamohammadi M, Haine D, Kelton DF, Barkema HW, Hogeveen H, Keefe GP, Dufour S. 2018. Herd-level mastitis-associated costs on Canadian dairy farms. Front Vet Sci. 5:100. doi: 10.3389/fvets.2018.00100
- Ali AKA, Shook GE. 1980. An optimum transformation for somatic cell concentration in milk. J Dairy Sci. 63:487–490. doi: 10.3168/jds.S0022-0302(80)82959-6
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21:263–265. doi: 10.1093/bioinformatics/bth457
- Berger T, Togawa A, Duncan GS, Andrew JE, You-Ten A, Wakeham A, Hannah EHF, Carol CC, Tak WM. 2005. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemiareperfusion injury. Proc Natl Acad Sci USA. 106:1834–1839.
- Breyne K, Steenbrugge J, Demeyere K, Vanden Berghe T, Meyer E. 2017. Preconditioning with lipopolysaccharide or lipoteichoic acid protects against Staphylococcus aureus mammary infection in mice. Front Immunol. 8:833. doi: 10.3389/fimmu.2017.00833
- Devireddy LR, Gazin C, Zhu X, Green MR. 2005. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 123:1293–1305. doi: 10.1016/j.cell.2005.10.027
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 432:917–921. doi: 10.1038/nature03104
- Flower DR. 1996. The lipocalin protein family: structure and function. Biochem J. 318:1–14. doi: 10.1042/bj3180001
- Holmberg M, Fikse WF, Andersson-Eklund L, Artursson K, Lundén A. 2012. Genetic analyses of pathogen-specific mastitis. J Anim Breed Genet. 129:129–137. doi: 10.1111/j.1439-0388.2011.00945.x
- Jensen K, Günther J, Talbot R, Petzl W, Zerbe H, Schuberth HJ, Seyfert HM, Glass EJ. 2013. Escherichia coli- and Staphylococcus aureus-induced mastitis differentially modulate transcriptional responses in neighbouring uninfected bovine mammary gland quarters. BMC Genomics. 14:36. doi: 10.1186/1471-2164-14-36
- Kjeldsen L, Johnsen AH, Sengeløv H, Borregaard N. 1993. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J Biol Chem. 268:10425–10432.
- Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron a metal at the host–pathogen interface. Cellular Microbiol. 12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x
- Patrushev LI, Kovalenko TF. 2014. Functions of noncoding sequences in mammalian genomes. Biochemistry (Mosc). 79:1442–1469. doi: 10.1134/S0006297914130021
- Pighetti GM, Elliott AA. 2011. Gene polymorphisms: the keys for marker assisted selection and unraveling core regulatory pathways for mastitis resistance. J Mammary Gland Biol Neoplasia. 16:421–432. doi: 10.1007/s10911-011-9238-9
- Pokorska J, Kułaj D, Dusza M, Żychlińska-Buczek J, Makulska J. 2016. New rapid method of DNA isolation from milk somatic cells. Anim Biotechnol. 27:113–117. doi: 10.1080/10495398.2015.1116446
- Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 laying down specific hygiene rules for food of animal origin.
- Robinson KM, McHugh KJ, Mandalapu S, Clay ME, Lee B, Scheller EV, Enelow RI, Chan YR, Kolls JK, Alcorn JF. 2014. Influenza A virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J Infect Dis. 209:865–875. doi: 10.1093/infdis/jit527
- Schepers AJ, Lam TJGM, Schukken YH, Wilmink JBM, Hanekamp WBA. 1997. Estimation of variance components for somatic cell counts to determine thresholds for uninfected quarters. J Dairy Sci. 80:1833–1840. doi: 10.3168/jds.S0022-0302(97)76118-6
- Schmidt-Ott KM, Mori K, Li JY, Kalandadze A, Cohen DJ, Devarajan P, Barasch J. 2007. Dual action of neutrophil gelatinase-associated lipocalin. J Am Soc Nephrol. 18:407–413. doi: 10.1681/ASN.2006080882
- Sharma N, Singh NK, Bhadwal MS. 2011. Relationship of somatic cell count and mastitis: an overview. Asian Austral J Anim. 24:429–438. doi: 10.5713/ajas.2011.10233
- Sharma N, Sodhi SS, Kim JH, Ghosh M, Zhang JJ, Koo D-B, Kang M-J, Jeong DK. 2016. Molecular cloning of lipocalin-2 into a eukaryotic vector and its expression in bovine mammary epithelial cells as a potential treatment for bovine mastitis. Turk J Biol. 40:55–68. doi: 10.3906/biy-1501-69
- Shashidharamurthy R, Machiah D, Aitken JD, Putty K, Srinivasan G, Chassaing B, Parkos CA, Selvaraj P, Vijay-Kumar M. 2013. Differential role of lipocalin 2 during immune complex-mediated acute and chronic inflammation in mice. Arthritis Rheum. 65:1064–1073. doi: 10.1002/art.37840
- Smith KD. 2007. Iron metabolism at the host pathogen interface: lipocalin 2 and the pathogen-associated iroA gene cluster. Int J Biochem Cell Biol. 39:1776–1780. doi: 10.1016/j.biocel.2007.07.003
- Srinivasan G, Aitken JD, Zhang B, Carvalho FA, Chassaing B, Shashidharamurthy R, Borregaard N, Jones DP, Gewirtz AT, Vijay-Kumar M. 2012. Lipocalin 2 deficiency dysregulates iron homeostasis and exacerbates endotoxin-induced sepsis. J Immunol. 189:1911–1919. doi: 10.4049/jimmunol.1200892
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. 2007. Primer3plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74. doi: 10.1093/nar/gkm306
- Xiao X, Yeoh BS, Vijay-Kumar M. 2017. Lipocalin 2: an emerging player in iron homeostasis and inflammation. Annu Rev Nutr. 37:103–130. doi: 10.1146/annurev-nutr-071816-064559