ABSTRACT
The study was conducted to investigate the effect of different levels of protein supplements on fertility and assess the fertility gene (BMP15 and GDP9) in sheep of Bangladesh. Thirty six ewes fed two protein level (Treatment 1, T1 and treatment 2, T2) and no protein supplement ration (control, T0). Polymerase chain reactions (PCR) were performed from blood cell DNA following standard protocols. PCR products were sequenced using Sanger sequencing and MEGA6 software were used for molecular analysis. The conception rate of ewes was significantly higher (P < 0.05) in the treated group than the control. Gestation period and lambing interval were significantly higher (worse) in control (154.00 ± 0.57 and 186.00 ± 1.24 days) than treated groups. Birth weight of lamb ranged from 1.07 to 1.65 kg and higher birth weight was found in T2 than other groups. Sequence alignment of BMP15 and GDF9 gene in indigenous sheep showed 100% homology with other sheep breed. As no mutation was observed and apparently these genes have no effect on ewe fertility. However, nutritional supplements had a positive impact on reproductive performance. The phylogenetic inferences have close relations of Bangladeshi sheep with other breed elsewhere.
1. Introduction
In Bangladesh, the peoples are facing a nutritional deficiency due to unavailability of a sufficient animal protein. Mutton (sheep meat) can play an important role to meet up the deficient amount of animal protein with comparatively lower prices. However, farmers who keep a significant number of sheep that are not ideally productive due to poor genetic merit (Rahman et al. Citation2014), indiscriminate breeding, poor nutrition and management (Alam et al. Citation2006), seasonal fluctuations of feed resources and diseases.
Reproductive performance of farm animal is mainly dependent on nutritional conditions, where poor nutrition results reduced body weight and body condition, delays the onset of puberty, increases the post-partum onset of estrus, interferes with normal ovarian cyclicity by decreasing gonadotropin secretion and increases infertility (Legesse Citation2008). For the improvement of the productive and reproductive status of animals, it may require improved ration especially by adding protein supplements. Protein-rich feed enhances the reproductive performance of ewes, but should be quality protein. However, the quality of the protein in a feed depends on both the amino acid profile and the digestibility.
In case of ewe, nutrition affects not only maternal body weight gain during gestation, body condition, and reproductive performance (Hess et al. Citation2005) but also affects prenatal and postnatal growth and development of offspring (Zohara et al. Citation2014). Improved diet in the ewe increases ovulation rate and modifies the morphological and functional quality of the embryo recovered (Ruiz-Lozano Citation2003). Several genes (such as BMP15, GDF9 and BMPR-1B) which are related to ovulation rate and litter size, have been exerted their effect on fecundity in sheep (Barzegari et al. Citation2010; Javanmard et al. Citation2011). Among them, BMP15 gene is responsible for the development and maturation of the oocytes (Javanmard et al. Citation2011). On the other hand, autosomal GDF9 has an essential role in controlling follicular growth through its influence on the granulosa cell function (Barzegari et al. Citation2010; Javanmard et al. Citation2011).
The advances of modern technologies are concerned about the animal fertility traits and identification of a gene responsible for these fertility traits. Nowadays, DNA isolation and gene identification have been widely used in domestic animals, including sheep in order to identify the genes responsible litter size (Javanmard et al. Citation2011) and ovulation (Hanrahan et al. Citation2004; Barzegari et al. Citation2010). However, no depth study has been done with fertility and its related genes in sheep using protein supplements. This concept will open a new horizon for the improvement of the fertility traits and reproductive efficiency of sheep. This exploration will be subsidiary to realize the effects of feed supplementation (protein) on fertility traits, and the efficient result will be accomplished in selection and breeding programme. For this purpose, this study was carried out to know the effects of protein supplements on fertility traits, and to assess the effect of BMP15 and GDF9 gene in the fertility of Bangladeshi sheep.
2. Materials and methods
2.1. Animal, study area and experimental design
The research work was conducted from January 2017 to June 2018 at the field and Poultry Research and Training Centre (PRTC) laboratory of CVASU, Bangladesh following the animal ethics rule and the ethics committee of Chattogram Veterinary and Animal Sciences University (CVASU), (Memo no. –CVASU/Dir (R&E) EC/2015/1011, Date 27/12/2018).
The study was carried out on thirty-six indigenous sheep, selected from three locations, namely Bakolia (09), Hathazari sadar (16) and Modonhat (11) in the Chattogram district of Bangladesh by direct visit, observation, body condition score (BCS, it estimates the condition of muscling and fat development over and around the vertebrae, and in the loin region the score ranged from 1 to 5, chosen score was 2 to 3), healthy and normal clinical conditions (free from diseases and parasites). All the locations had a similar ambient temperature, humidity, sheep management protocols. The age of studied sheep ranged from one to five years and was categorized as 1 to 2 (n = 10), >2 to 3 (n = 15) and >3 to 5 (n = 11) years. The sheep are allocated randomly in each age group and all location comprises of the same age category of sheep. The three groups of sheep were managed under the semi-intensive system, kept in separate pens and fed individually. The sheep fed with formulated rations in each location separately and each location was considered as a treatment (control/Bakolia, treatment 1/Hathazari sadar and treatment 2/Modonhat) with an unequal number of replications in each treatment.
2.2. Experimental ration formulation
Three iso-caloric rations (12 MJ/kg DM ME) containing graded levels of protein were formulated (11.68% CP for control/To, 12.94% CP for treatment 1/T1 and 13.96% CP for treatment 2/T2) by using of conventional feedstuffs () and concentrate mixture was supplied (0.250 kg/day/sheep) to all groups of sheep. Protein-concentrate (PRO-PAK) was provided in both T1 and T2 groups, but there was no protein concentrate added in the ration of the control group. The sheep of the three groups were allowed to graze for 6 to 7 h per day on the available natural grassland. It was assumed that the pasture value of these grasses was similar in all locations, as all location has the same available local grasses like durba (Cynodon dactylon), mutha (Cyperus rotundus), puti (Panicum spp.), chora (Paspalum diatatum), painna (Heteropogon spp.), regrowth paddy, road side grasses.
Table 1. Feed ingredients and chemical composition of experimental ration.
2.3. Record keeping for the fertility traits of the ewe
Data were recorded regarding the fertility traits (conception rate (%), gestation period (days), lambing interval (days) of the studied ewe population. The litter size (number) and birth weight (kg) of flock lamb born were recorded during the whole experimental period.
2.4. Molecular study of fertility genes of sheep
For molecular investigations of fertility traits (litter size and ovulation), blood was used for understanding the underlying biological processes.
2.4.1. Blood collection and DNA extraction
Thirty-six blood samples from the three treatments (T0 = 09; T1 = 16; T2 = 11) were collected from the jugular vein of mature ewes using vacutainer tube containing 0.5M EDTA (pH = 8). Genomic DNA was extracted by the FAVORGEN BIOTECH CORP and FavorPrepTM blood genomic DNA extraction mini kit. DNA was then stored at −20°C in a refrigerator before performing polymerase chain reaction (PCR). For genetic assessment 36 samples were tested for the presence of BMP15 and GDF9 genes, randomly from the studied sheep. Among all the samples, 32 (89%) samples were positive for both BMP15 and GDF9 genes, where typical amplicon sizes of the gene products measured by PCR and 4 sample were negative due to damage.
2.4.2. Primer designing and polymerase chain reaction (PCR) and sequencing
Primers for PCR of GDF9 and BMP15 genes were selected from published works (Hanrahan et al. Citation2004; Maitra et al. Citation2016) (). Polymerase chain reaction (PCR) was carried out with a standard protocol. The PCR products were visualized following electrophoresis through 1.8% ethidium bromide stained agarose gel and the fragments were photographed under gel documentation unit, and their sizes were estimated using a 1k bp DNA ladder (Fermentas International, Inc.).
Table 2. General characteristics of investigated genes (BMP15 and GDF9) (Hanrahan et al. Citation2004; Maitra et al. Citation2016).
The PCR product was cleaned with two μl of ExoSAP-IT (enzyme: ExoASP-IT) per sample. Then, it was incubated at 37°C for 15 min for degrading the remaining primers and nucleotides. Finally, inactivation of the ExoSAP-IT enzymatic reaction was done by incubating at 80°C for 15 min. Out of 32 positive sample 8 best purified PCR products ((T0 = 02; T1 = 03; T2 = 03), irrespective of the age of ewes were Sanger-sequenced with big dye terminator v3.1 sequencing kit and a 3730xl automated sequencer (Applied Biosystems, Foster City, CA, USA). The Nucleotide sequences were then determined on both strands of PCR amplification products at the Macrogen sequencing facility (Macrogen Inc., Seoul, Korea) using the ABI PRISM 3730XL Analyzer (96 capillary type).
2.5. Data analysis
The recorded data on different fertility traits (weight at conception (kg), conception rate (%), gestation period (days), lambing interval (days), litter size (no) and birth weight (kg)) of sheep were analyzed following randomized block design (RBD) using the Proc GLM of SAS (SAS Citation2008). The mean value was compared using the least significant different test (LSD) (Steel et al. Citation1997). The nucleotide sequences were analyzed to determine the sequence alignment, and the phylogenic tree was created using the neighbour-joining method by MEGA6 software (Tamura et al. Citation2013).
3. Results and discussion
3.1. Fertility traits of ewe’s
The various fertility traits of indigenous ewes of Bangladesh in consideration of treatment and age group are presented in . The conception rate (83.33%) in ewes in treatment group 2 was superior compared to other groups and differed significantly between treatments and control group, and this finding was agreed strongly with the findings of Zohara et al. (Citation2014). However, among the age group of ewes no differences were found in treatments for the conception rate. Conception rate was found to be superior with the increases of protein supplements and age of the sheep, this finding was similar to the other researcher (Kabir et al. Citation2004; Aktaş et al. Citation2015) but differed with the findings of Fukui et al. (Citation2010), they stated that old aged ewes had a less conception rate than 1 to 2 years aged ewes.
Table 3. Mean ± standard error of fertility traits of indigenous ewes of Bangladesh.
The gestation period in studied ewes varied from 150 to 155.50 days and was significantly lower (better) in the treated group than the control group. Nutrition, especially protein supplements, had a positive impact on pregnancy in ewes this also reported by Al- Haboby et al. (Citation1999) and Zohara et al. (Citation2014). No differences were observed among age groups in treatments for a gestation period of ewes. The gestation length was significantly higher in the control group compared with treated groups, which was similar to the findings of Zohara et al. (Citation2014) and inconsistent with Sultana et al. (Citation2011), who observed that gestation length was longer in the treatment groups than the control groups.
The lambing interval in the control group was 186.00 ± 1.24 days, which was significantly different (P < 0.05) from the treatment 2 group. This finding indicated that the higher protein supplement group was better than another group and this outcome was similar with Hassan and Talukder (Citation2012) but did not agree with the findings of Bhuiyan (Citation2014) and Poonia (Citation2008). Therefore, to minimize the lambing interval, post lambing estrus and service per conception should be minimized. Through dietary intervention and management minimizes the lambing interval in this study, which might be helpful to improve sheep production. On the other hand, the lambing interval significantly differed among age groups in treatment, which were supported by Aguirre Riofrio et al. (Citation2016).
The litter size was not significantly different between treatment and age group within a treatment (). The nutrition and age had no effect on the litter size of ewes, which were similar to the outcomes of Banerjee (Citation2008). However, the findings of the study disagreed with the results of Al- Haboby et al. (Citation1999), Sultana et al. (Citation2011) and Aktaş et al. (Citation2015), who showed that litter size was more in the protein supplement group than the control group (no supplements of protein).
The birth weight of lamb ranged from 1.07 to 1.65 kg and differed significantly in treatment. The birth weight of lambs in treatment 2 groups was 1.48 ± 0.07 kg, which was significantly higher compared to the other two groups. The mature aged (>3 years) sheep gave higher weighted lamb than other aged group ewes. The higher birth weight of lambs in treatment groups was followed the results of Kabir et al. (Citation2004) and Zohara et al. (Citation2014), they observed that the birth weight of lambs in the case of protein treated groups was higher than the control group. However, the current findings of birth weight were not supported by the findings of Hassan and Talukder (Citation2012), who observed, there was no relation of birth weight of lamb with feed treated groups. Aktaş et al. (Citation2015) showed that, age has a positive impact on birth weight. The amount and type of feed provided to pregnant ewe have influences on birth weight of lamb. From this study it can be seen that 13.96% CP supplement feed results useful for the reproductive performance of sheep.
3.2. PCR product and nucleotide sequences of BMP15 and GDF9 gene
The typical amplicon sizes of the BMP15 and GDF9 gene products from the positive samples measured by PCR are also shown in and (a,b), respectively. The sequences of the GDF9 gene are shown in and (a), and BMP15 gene fragment is shown in and (b), respectively. The sequence of the gene fragments was aligned with the reference gene sequences (from NCBI) of other sheep breeds (Pelibuey sheep, Norwegian white sheep) and observed a close similarity with their sequences. The sequences of BMP15 gene revealed 100% homology (NCBI accession no: KT853038.1) with the sequence of Pelibuey sheep breed (Argüello-Hernández et al. Citation2014), and 99% similar to that of Balochi breed (Nawaz et al. Citation2013) and Lohi breed of sheep (NCBI accession no: JN655672.1, JN655671.1) respectively. The sequences of the GDF9 gene fragment were 100% identical with the sequence of Norwegian white sheep (Våge et al. Citation2013) and Pelibuey sheep (Argüello-Hernández et al. Citation2014) (NCBI accession no: HE866499.1, KT853039.1), respectively. However, it showed 95% homology with that of Suffolk × Awassi crossbred (NCBI accession no: KT357485.1) and Awassi breed (NCBI accession no: KT357483.1). There was no mutation observed in BMP15 and GDF9 genes compared with wild type sheep in both control and protein supplemented treatment groups of indigenous sheep of Bangladesh and it does not affect the ewe fertility. Even though the presence of mutations in GDF9 and BMP15 has been reported that accountable for high prolificacy in sheep (Galloway et al. Citation2000), none of these significant mutations were detected in ewes in the current study. These findings indicate that the indigenous ewes of Bangladesh are genetically uniform irrespective to age groups for these two genes.
Figure 1. (a) PCR products of BMP15 gene 575 bp amplicon size; (b) PCR products of BMP15 gene 575 bp amplicon size.
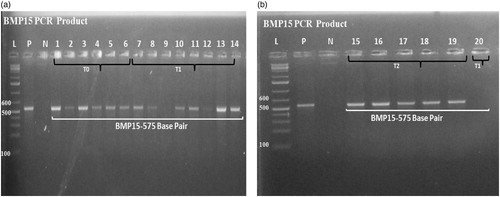
Figure 2. (a) PCR products of GDF9 gene 462 bp amplicon sizes; (b) PCR products of GDF9 gene 462 bp amplicon sizes.
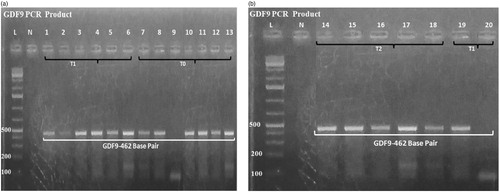
Figure 3. (a) The 100% identity sequence of OvisariesGDF9 genes (sequence ID: HE866499.1); (b) The 100% identity sequence of OvisariesBMP15 gene (sequence ID: KT853038.1).
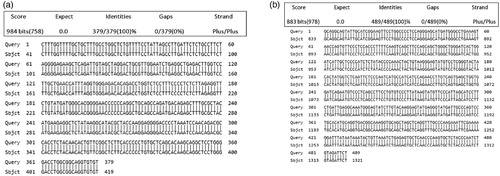
Figure 4. (a) Nucleotide sequences alignment of GDF9 gene (Control = G1 DNA_GDF9 & G2 DNA_GDF9; Treatment 1 = G3 DNA_GDF9, G4 DNA_GDF9 & G5 DNA_GDF9; and Treatment 2 = G6 DNA_GDF9, G7 DNA_GDF9 & G9 DNA_GDF9) (b) Nucleotide sequences alignment of BMP15 gene (Control = B1DNA_BMP15; Treatment 1 = B4DNA_BMP15 & B5DNA_BMP15; and Treatment 2 = B7 DNA_BMP15)
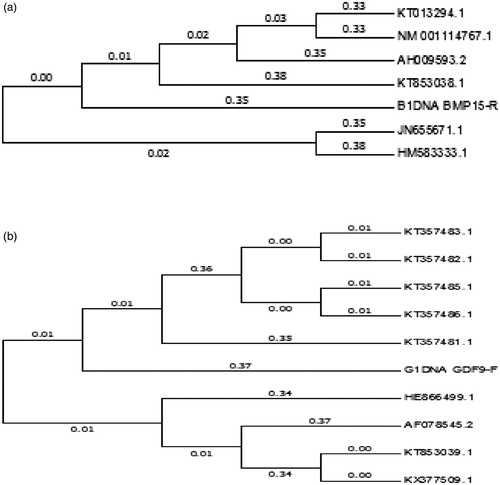
3.3. Evolutionary relationships (phylogenetic tree) taxa of BMP15 and GDF9 gene
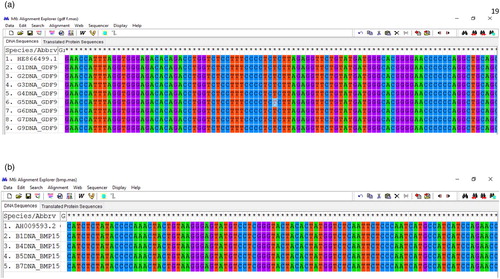
The phylogenetic tree for the study of evolutionary relationships taxa of BMP15 and GDF9 gene is shown in (a,b), respectively. The BMP15 gene, sample sequence (B1DNA BMPR15-R) are very close to the sequences of Pelibuey sheep (NCBI accession no: KT357481.1) (Argüello-Hernández et al. Citation2014) which are commonly found in Mexico, the Caribbean and South America than the sequences of Indian sheep (NCBI accession no: HM583333.1). For the GDF9 gene, the sample sequence (G1DNA GDF9F) is closer to the sequence of Egyptian Rahmani sheep (NCBI accession no: KT357481.1) than the sequence of Egyptian crossbred (Suffolk × Awassi) (NCBI accession no: KT357485.1) and Pelibuey sheep (NCBI accession no: KT853039.1) (Argüello-Hernández et al. Citation2014). The findings were similar to the phylogeny results of previous studies (for example, Bibinu et al. Citation2016) of BMP15 gene, who reported a similar clustering of sequences among the various breeds, although there was some intermingling process between the species. The phylogenetic relationship of these two investigated genes indicated that, the indigenous sheep of Bangladesh is similar to the sheep of Mexico, North America and Egypt. This might be due to similar environmental conditions and ancestor relatedness, which is the effect of an evolutionary process of these sheep.
Figure 5. (a) Phylogenetic tree drawn based on nucleotide sequences of the BMP15gene Legends: KT013294.1 = Ovis aries breed Lori Bakhtiari, NM001114767.1 = Ovis aries bone morphogenetic protein 15 (BMP15), mRNA,AH009593.2 = Ovis aries chromosome X growth differentiation factor 9B precursor,gene, complete cds, KT853038.1 = Ovis aries breed Pelibuey bone morphogenetic protein 15 (BMP15)mRNA, complete cds,B1DNA BMP15-R = Indigenous local sheep of Bangladesh,JN655671.1 = Ovis aries breed Lohi bone morphogenetic protein 15 (BMP15) gene,complete cds, HM583333.1 = Ovis aries isolate MS11R bone morphogenetic protein 15 (BMP15)gene, exon 2 and partial cds. (b) Phylogenetic tree drawn based on nucleotide sequences of the GDF9 gene Legends: KT357483.1 = Ovis aries breed Awassi (GDF-9)gene, partial cds, KT357482.1 = Ovis aries breed Barki(GDF-9)gene, partial cds, KT357485.1 = Ovis aries breed Suffolk X Awassi (GDF-9) gene, partial cds, KT357486.1 = Ovis aries breed Ossimi(GDF-9)gene, partial cds, KT357481.1 = Ovis aries breed Rahmani (GDF-9)gene, partial cds, G1DNAGD-F = Indigenous local sheep of Bangladesh, HE866499.1 = Ovis aries GDF9 gene, breed Norwegian white sheep, AF078545.2 = Ovis aries GDF-9 gene, complete cds, KT853039.1 = Ovis aries breed Pelibuey, KX377509.1 = Ovis aries breed Mehraban (GDF9)gene, exon 1 and partial cds.
4. Conclusion
The study showed that the protein-rich feed has a positive impact on the reproductive performance of ewes. The sequence alignment of BMP15 and GDF9 in the feeding groups has not shown mutation and it does not affect the ewe fertility. It is recommended that 13.96% CP supplement feed can be recommended for the grazing sheep for high fertility. The phylogenetic tree of BMP15 and GDP9 genes of indigenous sheep of Bangladesh indicated that these sheep has a genetic relationship with other indigenous breeds of sheep elsewhere. There were some limitations in this study, such as the few numbers of ewes in each treatment and less number of PCR products were sequenced per gene, which might result in no mutations. Confidence that the correct treatment can be chosen if additional data were to be collected and added in the current study. However, the genomic thoughts of BMP15 and GDF9 genes will be helpful to conserve the genetic constituents of indigenous sheep and can be applied for the genetic improvement of sheep.
Acknowledgements
The authors have expressed their sincere gratitude to the University Grants Commission (UGC) of Bangladesh and Chittagong Veterinary and Animal Sciences University for providing funds for implementing this research. The authors are immensely grateful to the farmers for helping them during experimental work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Aguirre Riofrio EL, Ferraz JBS, Mattos EC. 2016. Influence of non-genetic factors on growth and reproductive traits of sheep Santa Inês in extensive systems. Livestock Res Rural Dev. 28(7):Article #121 [accessed 2019 July 3]. http://www.lrrd.org/lrrd28/7/agui28121.html.
- Aktaş AH, Dursun Ş, Dŏgan Ş, Kiyma Z, Demirci U, Halıcı I. 2015. Effects of ewe live weight and age on reproductive performance, lamb growth, and survival in central anatolian merino sheep. Arch Anim Breed. 58:451–459. doi:10.5194/aab-58-451-2015.
- Alam MGS, Ul-Azam MS, Khan MJ. 2006. Supplementation with urea and molasses and body weight, milk yield and onset of ovarian cyclicity in cows. J Reprod Develop. 52:529–535.
- Al- Haboby AH, Salmana T, Abdul Kareem A. 1999. Influence of protein supplemetation on reproductive traits of Awassi sheep grazing cereal stubble. Sm Rumin Res. 34:33–40.
- Argüello-Hernández HJ, Cortez-Romero C, Rojas-Martínez RI, Segura-León OL, Herrera-Haro JG, Salazar-Ortiz J, Gallegos-Sanchez J. 2014. Origin, history and current situation of pelibuey sheep in Mexico. Trop Subtrop Agroecosys. 20:429–439.
- Banerjee R. 2008. Conservation and in situ development of a prolific indigenous sheep in the Sundarban and Sagar Island [PhD thesis]. Koltata: University of Calcutta [accessed 2018 April 26]. http://www.caluniv.ac.in.
- Barzegari A, Atashpa S, Ghabili K, Nemati Z, Rustaei M, Azarbaijani R. 2010. Polymorphisms in GDF9 and BMP15 associated with fertility and ovulation rate in Moghani and Ghezel sheep in Iran. Reprod Domest Anim. 45:666–669.
- Bhuiyan AKFH. 2014. Farm animal genetic resources in Bangladesh: diversity, conservation and management. Dhaka, Bangladesh: SAARC Agriculture Centre (SAC) Press, p. 174.
- Bibinu BS, Yakubu A, Ugbo SB, Dim NI 2016. Computational molecular analysis of the sequences of BMP15 gene of ruminants and non-ruminants. Open J Genet. 6:39.
- Fukui Y, Kohno H, Okabe K, Katsuki S, Yoshizawa M, Togari T, Watanabe H. 2010. Factors affecting the fertility of ewes after intrauterine insemination with frozen-thawed semen during the non-breeding season. J Reprod Dev. 56:460–466.
- Galloway SM, Mcnatty KP, Cambridge LM. 2000. Mutations in anoocyte-derive growth differentiation factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensetive manner. Nature Genet. 25:279–283.
- Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway SM. 2004. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol Reprod. 70:900–909.
- Hassan M, Talukder M. 2012. Comparative performance of different regional native sheep in Bangladesh. Bangladesh Veter. 28:85–95.
- Hess B, Lake S, Scholljegerdes E, Weston T, Nayigihugu V, Molle J, Moss G. 2005. Nutritional controls of beef cow reproduction. J Anim Sci. 83:E90–E106.
- Javanmard A, Azadzadeh N, Esmailizadeh AK. 2011. Mutations in bone morphogenetic protein 15 and growth differentiation factor 9 genes are associated with increased litter size in fat-tailed sheep breeds. Vet Res Commun. 35:157–167.
- Kabir F, Sultana M, Shahjalal M, Khan M, Alam M. 2004. Effect of protein supplementation on growth performance in female goats and sheep under grazing condition. Pak J Biol Sci. 3:237–239.
- Legesse G. 2008. Productive and economic performance of small ruminants in two production systems of the highlands of Ethiopia. Cuvillier Verlag. 14:12–13.
- Maitra A, Sharma R, Ahlawat S, Borana K, Tantia M. 2016. Fecundity gene SNPs as informative markers for assessment of Indian goat genetic architecture. Indian J Anim Sci. 50:349–356.
- Nawaz A, Masroor K, Babar E, Nadeem A, Hussain T, Bilal F, Javed K, Muhammad K. 2013. Identification of molecular markers in bone morphogenetic protien15 (BMP15) gene of Balochi sheep. Pak J Zool. 45:1351–1357.
- Poonia J. 2008. Reproductive performance of Munjal sheep. Indian J Small Rumin. 14:121–123.
- Rahman S, Begum IA, Alam MJ. 2014. Livestock in Bangladesh: distribution, growth, performance and potential. Livest Res Rural Devel. 26:233–238.
- Ruiz-Lozano JM. 2003. Arbuscular mycorrhizal symbiosis and alleviation of osmotic stress. New perspectives for molecular studies. Mycorrhiza. 13:309–317.
- SAS, Users guide statistics. 2008. Institute, Inc. Cary, North Carolina. Sign, 2, 1.
- Steel RGD, Torrie JH, Dickey DA. 1997. Principal and procedure of statistics- A biometrical approach. NewYork and London: Mc Graw-Hill, p. 39–177.
- Sultana N, Hassan N, Ershaduzzaman M, Talukder M, Iqbal A. 2011. Effect of intensive and semi-intensive feeding system on productive and reproductive performances of native sheep. J Sci Res. 3:693–698.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Våge DI, Husdal M, Kent MP, Klemetsdal G, Boman IA. 2013. A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep. BMC Genomics. 14:1.
- Zohara BF, Azizunnesa A, Islam M, Alam M, Bari FY. 2014. Exfoliative vaginal cytology and serum progesterone during the estrous cycle of indigenous ewes in Bangladesh. J. Embryo Transf. 29:183–188.
