ABSTRACT
As a member of the insulin-like growth factor mRNA binding proteins (IGFBPs) family, IGF2BP3 plays important role in skeletal muscle development by regulating IGF2, which has been considered a key gene related to growth and reproductive traits in many agricultural animal species. However, study on the goose IGF2BP3 gene is still scarce. In this study, the full-length cDNA sequence of goose IGF2BP3 gene was cloned and characterized. The goose IGF2BP3 cDNA is 1825 bp in length, containing an open reading frame (ORF) of 1755 bp that encodes a protein of 584 amino acids, which consists of two RNA-recognition motifs (RRMs) and four K-homologous (KH) domains. RT-qPCR analysis indicates that the IGF2BP3 mRNA expression level is the highest in ovary tissue, followed in hypothalamus, spleen, leg muscle and liver, and low level in breast muscle and muscular stomach of the adult Zhedong White goose. Meanwhile, 25 single nucleotide polymorphisms (SNPs) and 5 insertions/deletions (InDels) variants were identified in about 10,764 bp of goose IGF2BP3 genomic DNA sequence. These results serve as a foundation for further investigations of the function of IGF2BP3 gene in geese.
Introduction
The insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) belong to a conserved family of RNA binding, which contain three members including IGF2BP1, IGF2BP2 and IGF2BP3, and act in various important aspects of cell functions, such as cell polarization, migration, morphology, metabolism, proliferation and differentiation (Bell et al. Citation2013).
As a member of the IGF2BPs, IGF2BP3 is also known as IMP3/VICKZ3 and plays crucial role during cell developmental processes (Nielsen et al. Citation1999; Nassiry et al. Citation2005; Yisraeli Citation2005; Bell et al. Citation2013). Studies indicated that IGF2BP3 is a secreted protein that can bind to insulin-like growth factor 2 (IGF2), regulate IGF2 localization and play important roles in cell proliferation and migration (Jones and Clemmons Citation1995; Nielsen et al. Citation2002; Bell et al. Citation2013). Additionally, IGF2 is an important growth factor that can promote cell development and growth. It can bind to the two types of cell surface receptors known as IGF1 receptor and IGF2 receptor, stimulate cell proliferation and inhibit apoptosis (Bergman et al. Citation2013). Meanwhile, IGF2 gene is well known to be located within the linkage regions and has been considered as a positional candidate gene of growth traits in many animal species (Sewalem et al. Citation2002; McElroy et al. Citation2006; Wang et al. Citation2005). Therefore, IGF2BP3-mediated regulation of IGF2 function may play key role in animal muscle development (Lin et al. Citation2017).
In recent years, a series of studies have focused on the function of IGF2BP3 gene in different species. In humans, a study reported that IGF2BP3 as an RNA N6-methyladenosine reader can regulate cell cycle and angiogenesis in colon cancer, and it also had been associated the most with distinct cancer types (Hammer et al. Citation2005; Lederer et al. Citation2014; Yang et al. Citation2020; Zhang et al. Citation2022). A study on chicken performed by Ye et al. (Citation2014) showed that IGF2BP3 gene participated in adipocytokine and insulin signalling pathways. Subsequently, Lin et al. (Citation2017) demonstrated that IGF2BP3 was a direct target gene of let-7b, which can regulate myoblast proliferation by inhibiting IGF2BP3 expression in dwarf and normal chickens. Furthermore, a study performed by Yang et al. (Citation2021) indicated that DNA methylation regulated the expression and function of IGF2BP3 by modulating SP1 binding in the skeletal muscle development of pigs.
The goose is a popular poultry species, and in the past two decades, the goose industry has become highly profitable across the globe. As an important meat poultry, its growth and development have always been focused, and increased muscle growth in geese is economically beneficial for the poultry industry. IGF2BP3 plays an important role in skeletal muscle development, but the information on goose IGF2BP3 gene is limited. In this study, we isolated and analysed the full-length coding sequence of the goose IGF2BP3 gene, investigated its expression pattern in various tissues and screened the genetic variants located in this gene.
Materials and methods
Animals and samples collection
Six healthy female adult (70 days) Zhedong White geese were provided by Wenjie goose breeding of Xiangshan Co. LTD, China. All geese had access to open ground with a pool for swimming and reared under commercial management conditions. Several tissue samples, including heart, liver, spleen, lung, kidney, breast muscle, leg muscle, brain, skin, muscular stomach, hypothalamus, pituitary and ovary tissues were collected immediately, frozen in liquid nitrogen, and then stored at −80 °C prior to RNA isolation. Tissues pooled from leg muscle and brain were used for cloning the full-length cDNA sequence of goose IGF2BP3 gene.
Blood samples were collected under the goose wing vein to the disposable blood collection tubes those including EDTA anticoagulant, stored at −20°C before genomic DNA isolation. Blood samples from the three Chinese native goose breeds were used to screen the genetic variants located in the genomic DNA sequence of the IGF2BP3 gene. The blood samples of Zhedong White goose population were provided by Zhedong White Goose Institute of Xiangshan County (Ningbo, China), Shitou goose from Shantou Baisha Research Institute of Original Species of Poultry and Stock (Shantou, China), Zi goose from Heilongjiang Animal Science Institute (Qiqihaer, China). All animal experiments were handled in compliance with the Law of the People’s Republic of China on Animal Protection, performed according to the protocols of the Shanghai Academy of Agricultural Sciences and the Institutional Animal Care and Use Committee (Approval ID: SAASXM0522005).
RNA extraction, DNA isolation and cDNA synthesis
Total RNA was extracted with Trizol Reagent (Invitrogen, USA) and treated with RNase-free DNaseI (TaKaRa, Dalian) to remove any contaminating genomic DNA. First-strand cDNA was synthesized using ReverTra Ace Kit (TaKaRa, Dalian) and the concentration of each sample was adjusted to 200–300 ng/µL, stored at −20°C for quantitative real-time PCR (qPCR) analysis.
Genomic DNA was isolated from the blood samples using AxyPrepTM blood Genomic DNA Miniprep Kit (AxyGEN, USA). DNA concentration and quality were measured with the spectrophotometer ND-1000 (Nano-Drop, USA), and the concentration of each sample was adjusted to 50–300 ng/µL and stored at −20°C.
Molecular cloning and sequence analysis of goose IGF2BP3 gene
Based on the conserved region of IGF2BP3 gene in chicken (ID: NM_001006359) and duck (ID: XM_038174133), two pairs of primers (BP3-F1/R1 and BP3-F2/R2) were designed to amplify the whole CDS sequence of goose IGF2BP3 gene (). The PCR program included denaturation for 5 min at 94°C, followed by 36 cycles of 30 s at 94°C, 45 s at annealing temperature, 90 s at 72°C, and an extension step of 5 min at 72°C. The purified PCR products were cloned into the PEASY-T1 vector (TransGen, China) and sequenced commercially (Sangon, China).
Table 1. Primers information used in this study.
The obtained cDNA sequences were matched using DNAMAN software. The open reading frame (ORF) and the amino acid sequences were deduced using SeqMan (DNAstar, USA). Sequence similarity searches in GenBank were performed by using the BLAST2.1 search tool (http://www.ncbi.nlm.nih.gov/blast).
Multiple sequence alignments and phylogenetic analysis of IGF2BP3 gene
Multiple alignments of the amino acid sequences were performed and edited using CLUSTALW (http://www.genome.jp/tools-bin/clustalw) and the ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) software. The animal species for which a complete amino acid sequence was available, including Anas platyrhynchos (XP_038030061), Gallus gallus (XP_025002963), Taeniopygia guttata (XP_002192437), Columba livia (XP_005510979), Falco peregrinus (XP_005237255), Calypte anna (XP_030300644), Falco cherrug (XP_005445862), Sus scrofa (XP_020919108), Bos taurus (NP_001179217) and Ovis aries (XP_004007841).
A phylogenetic tree was constructed based on the above amino acid sequence alignment by the neighbour-joining algorithm of the MEGA 5.0 software (Tamura et al. Citation2011). The reliability of the branching was tested by bootstrap re-sampling (1000 replicates). The evolutionary distances were computed using the Jones-Thornton-Taylor matrix-based method (Jones et al. Citation1992).
Tissue expression pattern analysis of goose IGF2BP3 mRNA
To detect the IGF2BP3 mRNA levels in different tissues of the female adult (70 days) Zhedong White geese, qPCR was performed with a 384-well C1000 TouchTM Thermal (Bio-Rad, USA) using TB Green Premix Ex Taq II (Tli RNaseH Plus) (Bio-Rad, USA). IGF2BP3-QF/QR (ID: ON908307) and β-actin-F/R (ID: M26111.1) primers were designed and used for qPCR analysis (). The PCR program included a denaturation step of 2 min at 95°C, followed by 40 cycles of 5 s at 95°C, 30 s at 60°C. The specificity of qPCR primers for each gene was confirmed by melting curve analysis. Melting curve analysis parameters were 1 cycle at 95°C for 10 s, 1 cycle at 65°C for 5 s, and then the temperature was increased 0.5°C/cycle to 95°C while continuously monitoring fluorescence. The relative quantification of gene expression was replicated three times for each sample. The data were analysed with one-way ANOVA, followed by Tamhane’s T2 post hoc test. The results were expressed as the mean ± SEM. p < 0.05 was considered statistically significant, and p < 0.01 was considered very significant.
Genetic variants identification of goose IGF2BP3 gene
The three goose populations were applied to construct the DNA pooling. According to the genomic DNA sequence of Peking duck IGF2BP3 (NC_040047), 14 pairs of primers (BP3-SF1/SR1~BP3-SF14/SR14, ) were synthesized to identify the genetic variants in the regions of the whole CDS, several introns and partial 3’-untranslated region (UTR) of goose IGF2BP3 gene. PCR products were amplified from the DNA pools and sequenced directly by the SangonBiotech (Shanghai, China). The obtained sequences were aligned by SeqMan of DNASTAR software.
Results
cDNA sequence analysis of goose IGF2BP3 gene
Using sequence matching techniques, a cDNA sequence covering the whole coding sequence was obtained from the tissue mixture of goose. The IGF2BP3 cDNA (ID: ON908307) consists of 1825 nucleotides, containing an ORF of 1755 bp, a 5’-UTR of 23 bp, and a 3’-UTR of 47 bp (). The goose IGF2BP3 cDNA shares high similarity to its counterpart in Anas platyrhynchos (XM_038174135, 98.90%), Gallus gallus (XM_001006359, 96.64%), Falco cherrug (XM_005445805, 97.04%) and Coturnix Japonica (XM_015853829, 95.76%). Bioinformatics analysis indicated that the goose IGF2BP3 encodes a protein of 584 amino acids (aa), which consists of six typical canonical RNA-binding domains, including two RNA recognition motifs (RRM) and four K-homology (KH) domains ().
Figure 1. Composite nucleotide and deduced amino acid sequences of the goose IGF2BP3 gene. A single open reading frame of 1755 bp is present, encoding a protein of 584 amino acids. The letters underlined and in bold indicate the start codon (ATG); the stop codon (TAA) is indicated with an asterisk; the two RRMs are shaded with light grey and dark grey, respectively; the four KH domains are indicated by a double underline, dotted line, bold underline and solid lines, respectively.

Multiple sequence alignments and evolutionary relationships of IGF2BP3
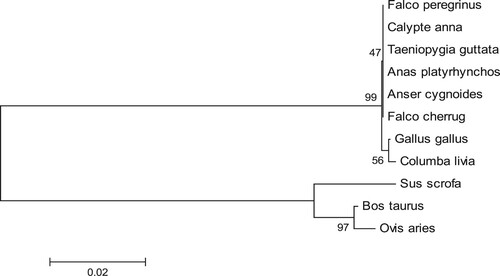
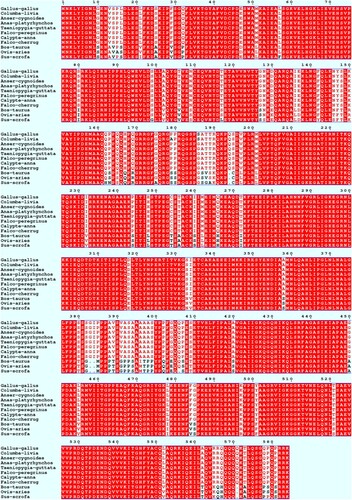
Multiple sequence alignment analysis revealed that the avian IGF2BP3 proteins were a little longer than those of the mammalian counterparts. Meanwhile, the IGF2BP3 proteins were highly conserved between the avian and mammalian species, and the IGF2BP3 proteins were completely conserved in Anser cygnoides, Anas platyrhynchos, Taeniopygia guttata, Falco peregrinus, Calypte anna and Falco cherrug ().
Figure 2. Multiple alignments of IGF2BP3 amino acid sequences among different species. Amino acids are presented in conventional single-letter code and numbered on the sequence. Amino acids that are identical among species are shown in white letters on a red background. Dashes indicate missing amino acids.
Based on the multiple sequence alignments, a phylogenetic tree was constructed using MEGA 5.0 software (). The dendrogram was clustered into two subgroups, the avian species including Anas platyrhynchos, Gallus gallus, Taeniopygia guttata, Coturnix japonica, Columba livia, Falco peregrinus, Calypte anna and Falco cherrug belonging to one group, and the mammal species including Sus scrofa, Bos Taurus and Ovis aries belonging to the other. The phylogenetic tree also displayed that the deduced goose IGF2BP3 protein was more closely related to the avian IGF2BP3, especially to that of Anas platyrhynchos, Taeniopygia guttata, Falco peregrinus, Calypte anna and Falco cherrug ().
Tissue expression pattern of goose IGF2BP3 gene
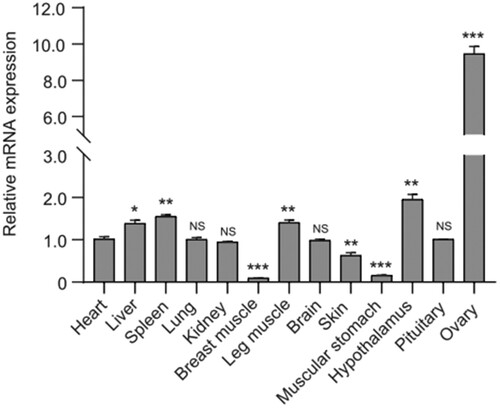
As shown in , qPCR analysis showed that the goose IGF2BP3 mRNA was differentially expressed in the detected tissues of the female adult (70 days) Zhedong White geese. The highest level of IGF2BP3 mRNA expression was observed in ovary tissue, and the levels in the hypothalamus, spleen, leg muscle and liver tissues were higher than that in heart, lung, kidney, brain, pituitary and skin, little expressed in breast muscle and muscular stomach tissues ().
Figure 4. Expression profile of IGF2BP3 mRNA in various tissues of the female adult Zhedong White geese. The relative expression level of IGF2BP3 mRNA was analysed with the Comparative Ct method, employing goose β-actin as the reference gene in each sample (n = 3); Values are displayed as the mean ± SEM. “*” shows significant differences (p < 0.05), “**” and “***” represent extremely significant differences (p < 0.01).
Genetic variation of goose IGF2BP3 gene
The sequences from the fourteen pairs of primers (BP3-SF1/SR1–BP3-SF14/SR14) were aligned by SeqMan of DNASTAR software. Sequence analysis displayed that no mutation was identified in the PCR products of the primer pairs BP3-SF6/SR6, BP3-SF7/SR7, BP3-SF12/SR12, BP3-SF13/SR13 and BP3-SF14/SR14. A total of 30 variations, including 25 SNPs and 5 InDels were identified in about 10,764 bp sequence from the products of other nine pairs of primers (). Eight mutations (T189C, T219C, C558G, T586C, T355C, A477G, T617G and G24A) located in the intron 2 region; one InDel (A100-) and one mutation (A235G) located in the intron 3, two InDels (T88– and CT94–) located in the intron 5, one synonymous mutation (A264G, Pro180Pro) located in the exon 6 region, seven mutations (C463G, C561T, C596T, G599C, C674A, C702T and C720T) located in the intron 6, one mutation (G554C) located in the intron 8, three mutations (T679C, C680T and G38A) and one InDel (TCAG53–-) located in the intron 10, G746A located in the intron 11, one InDel (AAT331–) and three mutations (A379G, C443T and A583G) located in the intron 12, respectively ().
Table 2. Polymorphisms detected in goose IGF2BP3 gene.
Discussion
The insulin-like growth factor-2 mRNA-binding proteins (IGF2BP1, IGF2BP2 and IGF2BP3) belong to a highly conserved family of RNA-binding, which carry six typical canonical RNA-binding domains, including two RRMs in their N-terminal part and four KH domains in the C-terminal region (Nielsen et al. Citation2001; Yaniv and Yisraeli Citation2002; Yisraeli Citation2005). In this study, we first cloned and characterized the goose IGF2BP3 cDNA sequence, which consists of an open reading frame of 1755 nucleotides with the capacity to encode a protein of 584 amino acids. Multiple sequence alignment implied that the number and sequences of IGF2BP3 amino acids were strongly conserved among the avain species. Meanwhile, the completely conserved IGF2BP3 amino acids sequences highlight it may act similar role on the Anser cygnoides, Anas platyrhynchos, Taeniopygia guttata, Falco peregrinus, Calypte anna and Falco cherrug. The sequence information from this study will facilitate further research on the function of the IGF2BP3 gene in geese.
The expression of eukaryotic genes is temporarily and multidimensional controlled (Shahsavari et al. Citation2021). Only a relatively small set of the entire genome is expressed in each type of tissue, and the expression of genes depends on the stage of development (Mohammadabadi et al. Citation2021). Meanwhile, the amount of gene products that are made in the same tissue as well as in other tissues that make up that product, regulates the expression of that gene (Direkvandi et al. Citation2020). One of the basic activities in domestic animals is the study of genes and proteins at the cellular or chromosomal level which are to economic traits (Mohamadipoor-Saadatabadi et al. Citation2021). Compared to the high expression level in the embryo, IGF2BP3 mRNA was reported to be expressed at negligible levels in adult mouse and human organs, but except reproductive tissues including ovaries, testis and brain tissues (Hammer et al. Citation2005). In Zebrafish, a high expression level also was detected in ovary tissue (Ren et al. Citation2020). In the present study, the highest IGF2BP3 mRNA expression level also was detected in the ovary tissue of adult Zhedong White goose, which implies that the goose IGF2BP3 may act important roles in ovarian follicle development, and be a strong performance target for goose reproductive traits. Therefore, further work will be necessary to confirm its influence on reproductive traits.
The determination of gene polymorphism is important for farm animals breeding to define genotypes of animals and their associations with productive, reproductive and economic traits (Yaripour et al. Citation2017). IGF2BP3 is a translational activator of IGF2, which was reported as a key gene located in the linkage regions affecting growth, carcass and reproductive traits of many agricultural animal species, such as pig, cattle, chicken, Muscovy duck and goose (Van Laere et al. Citation2003; Wang et al. Citation2005; Tang et al. Citation2010; Huang et al. Citation2014; Wang et al. Citation2021). Meanwhile, the whole-genome analysis revealed the association between IGF2BP3 gene and birth weight of humans (Warrington et al. Citation2019). Recently, Yang et al. (Citation2021) reported that IGF2BP3 might be considered as a candidate gene in breeding for meat production traits in the pig industry. In this study, 19 variants located in the intronic regions, only one synonymous mutation (A264G, Pro180pro) located in the exonic region, which reveals that the IGF2BP3 amino acid sequence is highly conserved in geese. Although these variants do not directly change the amino acid structure of the protein, they may affect the gene function by affecting translation with codon bias, changing the stability of the mRNA or controlling transcription of the gene (Zhang et al. Citation2005; Van Laere et al. Citation2003; Kimchi-Sarfaty et al. Citation2007; Hou et al. Citation2010). The potential effects of the large number of variations on goose economic traits will be investigated in our following work.
In this study, we first cloned and characterized the whole CDS sequence of the goose IGF2BP3 gene, and found the IGF2BP3 mRNA was highly expressed in the ovary tissues of the female adult Zhedong White geese, and identified 25 SNPs and 5 InDels variants in three native goose populations. All the present findings will provide important resources for research on the functions of the goose IGF2BP3 gene.
Acknowledgements
We thank the Special Project of Modern Agriculture (2021Z131), the China Agriculture Research System of MOF and MARA (CARS-42-7) and the SAAS Program for Excellent Research Team (2022-021) for supporting this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. 2013. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 70(15):2657–2675. doi:10.1007/s00018-012-1186-z.
- Bergman D, Halje M, Nordin M, Engstrom W. 2013. Insulin-like growth factor 2 in development and disease: a mini-review. Gerontology. 59(3):240–249. doi:10.1159/000343995.
- Direkvandi E, Mohammadabadi T, Salem A. 2020. Effect of microbial feed additives on growth performance, microbial protein synthesis, and rumen microbial population in growing lambs. Transl Anim Sci. 4(4):txaa203. doi:10.1093/tas/txaa203.
- Hammer NA, Hansen TV, Byskov AG, Rajpert-De Meyts E, Grøndahl ML, Bredkjaer HE, Wewer UM, Christiansen J, Nielsen FC. 2005. Expression of IGF-II mRNA-binding proteins (IMPs) in gonads and testicular cancer. Reproduction. 130(2):203–212. doi:10.1530/rep.1.00664.
- Hou G, Wang D, Guan S, Zeng H, Huang X, Ma Y. 2010. Associated analysis of single nucleotide polymorphisms of IGF2 gene's exon 8 with growth traits in Wuzhishan pig. Mol Bio Rep. 37(1):497–500. doi:10.1007/s11033-009-9681-5.
- Huang YZ, Zhan ZY, Li XY, Wu SR, Sun YJ, Xue J, Lan XY, Lei CZ, Zhang CL, Jia YT, Chen H. 2014. SNP and haplotype analysis reveal IGF2 variants associated with growth traits in Chinese Qinchuan cattle. Mol Biol Rep. 41(2):591–598. doi:10.1007/s11033-013-2896-5.
- Jones JI, Clemmons DR. 1995. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 16(1):3–34. doi:10.1210/edrv-16-1-3.
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8(3):275–282. doi:10.1093/bioinformatics/8.3.275.
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 315(5811):525–528. doi:10.1126/science.1135308.
- Lederer M, Bley N, Schleifer C, Hüttelmaier S. 2014. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 29:3–12. doi:10.1016/j.semcancer.2014.07.006.
- Lin S, Luo W, Ye Y, Bekele EJ, Nie Q, Li Y, Zhang X. 2017. Let-7b regulates myoblast proliferation by inhibiting IGF2BP3 expression in dwarf and normal chicken. Front Physiol. 8:477. doi:10.3389/fphys.2017.00477.
- McElroy JP, Kim JJ, Harry DE, Brown SR, Dekkers JC, Lamont SJ. 2006. Identification of trait loci affecting white meat percentage and other growth and carcass traits in commercial broiler chickens. Poult Sci. 85(4):593–605. doi:10.1093/ps/85.4.593.
- Mohamadipoor-Saadatabadi L, Mohammadabadi M, Amiri-Ghanatsaman Z, Babenko O, Stavetska R, Kalashnik O, Kucher D, Kochuk-Yashchenko O, Asadollahpour-Nanaei H. 2021. Signature selection analysis reveals candidate genes associated with production traits in Iranian sheep breeds. BMC Vet Res. 17(1):369. doi:10.1186/s12917-021-03077-4.
- Mohammadabadi M, Masoudzadeh SH, Khezri A, Kalashnyk O, Stavetska RV, Klopenko NI, Oleshko VP, Tkachenko SV. 2021. Fennel (Foeniculum vulgare) seed powder increases delta-like non-canonical Notch Ligand 1 gene expression in testis, liver, and humeral muscle tissues of growing lambs. Heliyon. 7(12):e08542. doi:10.1016/j.heliyon.2021.e08542.
- Nassiry MR, Shahroodi FE, Mosafer J, Mohammadi A, Manshad E, Ghazanfari S, Mohammad Abadi MR, Sulimova GE. 2005. Analysis and frequency of bovine lymphocyte antigen (BoLA-DRB3) alleles in Iranian Holstein cattle. Genetika. 41(6):817–822.
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 19(2):1262–1270. doi:10.1128/MCB.19.2.1262.
- Nielsen FC, Nielsen J, Christiansen J. 2001. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl. 234:93–99.
- Nielsen FC, Nielsen J, Kristensen MA, Koch G, Christiansen J. 2002. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J Cell Sci. 115(10):2087–2097. doi:10.1242/jcs.115.10.2087.
- Ren F, Lin Q, Gong G, Du X, Dan H, Qin W, Miao R, Xiong Y, Xiao R, Li X, et al. 2020. Igf2bp3 maintains maternal RNA stability and ensures early embryo development in zebrafish. Commun Biolo. 3(1):94. doi:10.1038/s42003-020-0827-2.
- Sewalem A, Morrice DM, Law A, Windsor D, Haley CS, Ikeobi CO, Burt DW, Hocking PM. 2002. Mapping of quantitative trait loci for body weight at three, six, and nine weeks of age in a broiler layer cross. Poult Sci. 81(12):1775–1781. doi:10.1093/ps/81.12.1775.
- Shahsavari M, Mohammadabadi M, Khezri A, Asadi Fozi M, Babenko O, Kalashnyk O, Oleshko V, Kachenko S. 2021. Correlation between insulin-like growth factor 1 gene expression and fennel (Foeniculum vulgare) seed powder consumption in muscle of sheep. Anim Biotechnol. 1–11. doi:10.1080/10495398.2021.2000997.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. doi:10.1093/molbev/msr121.
- Tang S, Sun D, Ou J, Zhang Y, Xu G, Zhang Y. 2010. Evaluation of the IGFs (IGF1 and IGF2) genes as candidates for growth, body measurement, carcass, and reproduction traits in Beijing You and Silkie chickens. Anim Biotechnol. 21(2):104–113. doi:10.1080/10495390903328090.
- Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, et al. 2003. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 425(6960):832–836. doi:10.1038/nature02064.
- Wang C, Liu Y, Zhang Y, Yang Y, Wang X, Li G, Wang H, Gong S, Chen S, He D. 2021. Insulin-like growth factor 2 (IGF2) gene: molecular characterization, expression analysis and association of polymorphisms with egg production traits in goose (Anser cygnoides). Anim Biotechnol. 1–9. doi:10.1080/10495398.2021.2015603.
- Wang G, Yan B, Deng X, Li C, Hu X, Li N. 2005. Insulin-like growth factor 2 as a candidate gene influencing growth and carcass traits and its bialleleic expression in chicken. Sci China C Life Sci. 48(2):187–194. doi:10.1007/BF02879672.
- Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B, … Freathy RM. 2019. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 51(5):804–814. doi:10.1038/s41588-019-0403-1.
- Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. 2020. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res.: CR. 39(1):203. doi:10.1186/s13046-020-01714-8.
- Yang Y, Fan X, Yan J, Chen M, Zhu M, Tang Y, Liu S, Tang Z. 2021. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 49(3):1313–1329. doi:10.1093/nar/gkaa1203.
- Yaniv K, Yisraeli JK. 2002. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 287(1-2):49–54. doi:10.1016/s0378-1119(01)00866-6.
- Yaripour S, Esmaeili HR, Gholamhosseini A, Rezaei M, Sadeghi S. 2017. Assessment of genetic diversity of an endangered tooth-carp, Aphanius farsicus (Teleostei: Cyprinodontiformes: Cyprinodontidae) using microsatellite markers. Mol Biol Res Commun. 6(4):153–160. doi:10.22099/mbrc.2017.24404.1246.
- Ye Y, Lin S, Mu H, Tang X, Ou Y, Chen J, Ma Y, Li Y. 2014. Analysis of differentially expressed genes and signaling pathways related to intramuscular fat deposition in skeletal muscle of sex-linked dwarf chickens. Biomed Res Int. 2014:724274. doi:10.1155/2014/724274.
- Yisraeli JK. 2005. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 97(1):87–96. doi:10.1042/BC20040151.
- Zhang BZ, Lei MG, Deng CY, Xiong YH, Li FE. 2005. Association between PCR-RFLP polymorphism of the fifth intron in lipoprotein lipase gene and productive traits in Pig resource family. Asian-Australas J Anim Sci. 18(4):458–462.
- Zhang W, Liu L, Zhao S, Chen L, Wei Y, Chen W, Ge F. 2022. Research progress on RNA-binding proteins in breast cancer. Oncol Lett. 23(4):121. doi:10.3892/ol.2022.13241.