?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Previous studies reported that DDB1 and CUL4 associated factor 7 (DCAF7) is associated with craniofacial, muscle, fat and bone growth and development. Therefore, the objective of this study was to explore genetic variation in the DCAF7 gene and its potential association with number of thoracolumbar vertebrae and carcass traits in Dezhou donkeys. Seven single-nucleotide polymorphisms (SNPs) were detected by targeted sequencing and Sanger sequencing in the intron region of the DCAF7 gene in 406 Dezhou donkeys. The polymorphism at g.48991059 T > G was significantly associated with hide weight (P < 0.05). g.48978712 T > C, g.48985896 A > G, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T sites were associated with number of thoracic vertebrae (P < 0.05). Furthermore, linkage disequilibrium analysis showed that four of the seven SNPs of DCAF7 gene were strongly linked to each other and can be used as Tag SNP. Data analysis revealed that haplotype combination Hap2Hap4 (TCAGTTCC) had the highest length of thoracic vertebrae, it was significantly higher than that in haplotype combinations Hap1Hap2 (TTAAGTCC) and Hap1Hap3 (TCAGGTCT) (P < 0.05). Finally, the results of this study suggest that the polymorphisms of DCAF7 gene can be used as a molecular marker for donkey breeding.
Introduction
Dezhou donkey is one of large donkey breeds in China, with characteristics as tall and muscular with compact structure, good growth performance (Lai et al. Citation2020; Zhang et al. Citation2021). Donkey products have become more popular in recent years by its nutritional value, such as donkey meat (Li et al. Citation2021) and donkey milk (Li et al. Citation2022). However, as a special economic animal, the progress of donkey breeding is slowly because of the longer growth cycle and interval between generations, and lower litter size compare with pigs, cattle and sheep .
Economic traits such as body length, carcass weight are important indicators for breeding. Former research showed that the number of vertebrae was found to be one of the important factors within determining the body length (Li et al. Citation2022; Liu, Gao et al. Citation2022). Liu, Gao et al. (Citation2022) have studied that the number of thoracolumbar vertebrae had positive effect on the body length and carcass weight of donkeys. Dezhou donkeys have three types of thoracic vertebrae i.e. 17, 18, 19 and 2 lumbar vertebrae, 5 and 6 (Liu, Gao et al. Citation2022). In European commercial pig breeds, the body length increases by about 15 mm for each additional vertebra. In Asian Kazakh sheep, the carcass length of 20 thoracolumbar vertebrae increased by 2.22 ∼ 2.93 cm compared to that of 19 thoracolumbar vertebrae (Li et al. Citation2017). In Dezhou donkey, increased one thoracolumbar vertebrae could lead to an increase in body length about 3 cm, and in carcass weight about 6 kg (Liu, Gao et al. Citation2022). The heritability of pig vertebrae is estimated to be 0.60–0.62, however the heritability of donkey vertebrae has not been reported (Fan et al. Citation2013).
Multi-vertebrae trait is regulated by multiple genes. Several genes have been shown to be associated with the number of thoracolumbar vertebrae in domestic animals. The TGFβ3 gene g.1051794747 locus is associated with the number of ribs and thoracolumbar vertebrae in pigs (Yue et al. Citation2018). Mutations in intron 4 of the ActRIIB gene are associated with the number of vertebrae in small-tailed cold sheep (Liu et al. Citation2010). The NR6A1 gene has previously been shown to play an important role in regulating the development of the thoracolumbar vertebrae in Dezhou donkeys (Liu, Gao et al. Citation2022). Bharambe et al. (Citation2020) found that DCAF7 was associated with the development of the thoracolumbar vertebrae. DCAF7 gene is an extremely important member of the DCAF gene family. Previous research has shown that DCAF7 gene is involved in the proliferation and differentiation of vertebrate somites via a synergistic activation of the Notch signalling pathway (Bharambe et al. Citation2020). In early embryonic development, the somites are the precursors of the spine and the total number of vertebrae depends on the number of mesodermal somites. Several research have revealed that the DCAF7 gene is involved in growth and development, including craniofacial development in human and zebrafish (Nissen et al. Citation2006; Wang et al. Citation2013; Leslie et al. Citation2016), adipocyte synthesis in human (Porstmann et al. Citation2008; Napolitano et al. Citation2020) and muscle development regulation in human and Drosophila (Morriss et al. Citation2013; Yu et al. Citation2019). Other studies in zebrafish showed that DCAF7 gene was essential to cartilage and craniofacial skeleton growth, especially the dorsal and abdominal cartilage (Alvarado et al. Citation2016). The DCAF7 gene is involved in the formation of the jaws of the platanna (Bonano et al. Citation2018). However, the effect of DCAF7 gene on the number of thoracolumbar vertebrae and carcass traits in donkeys has not been reported. Donkey DCAF7 gene is located on chromosome 13, contains eight exons and seven introns, encodes 342 amino acids in donkeys (Wang et al. Citation2020). Considering that the DCAF7 gene is reported to be associated with the proliferation and differentiation of vertebrate somites, therefore we hypothesized that DCAF7 may associated with skeletal development, the number of thoracolumbar vertebrae and carcass traits in Dezhou donkeys. The aim of this study was to investigate the polymorphism of the DCAF7 gene and its association with number of thoracolumbar vertebrae and carcass traits in Dezhou donkeys using targeted sequencing methods. This study could provide a foundation for breeding multi-thoracolumbar vertebrae donkeys.
Materials and methods
Ethics statement
The experimental animals and methods used in this study were approved by Animal Policy and Welfare Committee of Liaocheng University (No. LC2019-1). The care and use of laboratory animals is in full compliance with local animal welfare laws, guidelines and policies.
Animals and phenotypes
A total of 406 male Dezhou donkeys aged 22–24 months were investigated. All samples were collected at the slaughterhouse of Dezhou, Shandong Province during 2018–2021. All donkeys come from the same farm and are raised in the same feeding environment. The sampling season is in winter. Blood sample was collected from each Dezhou donkey using jugular vein blood collection and placed in an EDTA anticoagulated blood collection tube and stored at −80°C (Lai et al. Citation2020). Body height, body length and chest circumference were measured in accordance with the National Standard of the People's Republic of China ‘Dezhou Donkey’. Hide weight, carcass weight, number of lumbar vertebrae, number of thoracic vertebrae, length of lumbar vertebrae, length of thoracic vertebrae, total number of thoracic and lumbar vertebrae were measured after humanely slaughtered. Hide weight was determined immediately after slaughter and other carcass trait data were collected according to the method of Liu, Gao et al. (Citation2022).
DNA extraction
The 406 genomic DNA samples of Dezhou donkey were extracted from whole blood by TIANamp blood DNA Kit (Tiangen, Beijing, China), then detected DNA purity (OD260/OD280) using spectrophotometer (B500, Metash, China) and the quantity of extracted genomic DNA sample was detected by 1% agarose gel (Zheng et al. Citation2019).
SNP detection and genotyping
Qualified DNA samples were detected by the Molbreeding Biotechnology Co., Ltd. (Shijiazhuang, China) for targeted sequencing. The targeted sequencing method is a technique to achieve accurate genotype detection by high-depth resequencing of target genes. A total of 462 probes were used in the targeted sequencing, covering 98.01% of the DCAF7 gene with reference sequence as the DCAF7 gene (GenBank accession number NC_052189.1) in targeted sequencing.
SNPs validation
Based on the results of SNPs detected by targeted sequencing, the results of genotype frequencies less than 5% were removed. Primers were designed for SNPs with genotype frequencies greater than 5%. Based on the nucleotide sequence of the Dezhou donkey DCAF7 gene (GenBank accession number NC_052189.1), seven pairs of polymerase chain reaction (PCR) primers were designed to amplify seven mutation sites using primer premier software (version 5.0) () (Sun et al. Citation2012). One DNA sample per genotype was randomly selected for PCR amplification. A total of 25 μL of PCR amplification system, including 12.5 μL PCR Mix (Mei5bio, Beijing, China), 8.5 μL ddH2O, upstream primer 1 μl, downstream primer 1 μl, template 2 μl (Jin et al. Citation2019). The PCR procedure is shown in Table S1. The PCR products were sequenced by BGI Genomics Co., Ltd (Zheng et al. Citation2019). Sanger sequencing was used to validate the targeted sequencing results.
Table 1. Primer sequences, annealing temperature, and products size for Dezhou donkey DCAF7 gene.
Statistical analysis
Chromas software was used to read Sanger sequencing peak maps. The polymorphism information content (PIC), homozygosity (Ho), heterozygosities (He) and the effective number of alleles (Ne) were analysed using the GDIcall Online Calculator (http://www.msrcall.com/ Gdicall.aspx) (Lai et al. Citation2020). The haplotypes were constructed and the association between D’ (LD’ coefficient) and r2 (correlation coefficient) was calculated by the haploview software (Version 4.1. Daly lab at the Broad Institute Cambridge, USA) (Barrett et al. Citation2005). SPSS 26.0 software (IBM Statistics, Armonk, NY, USA) was used to analyse the correlation of the thoracolumbar vertebrae number and carcass traits with genotype and haplotype combinations of the mutation sites. All statistical tests were two-sided and a P < 0.05 was considered to indicate statistical significance (Erdenee et al. Citation2021). The linear analysis model can be written as
where Y is the individual phenotypic measurements, µ represents the mean for each trait, a represents the fixed factor genotype, e represents the random error. Least squares means with standard errors were used for the different genotypes and for the number of thoracolumbar vertebrae as well as the carcass traits. Age, sex and rearing environment were consistent, they were not included in the model. The different genotypes were considered as fixed effects, the random error as a random effect and the number of thoracolumbar vertebrae and carcass traits as the dependent variable (Yang et al. Citation2020). Multiple comparisons of the associations were based on Bonferroni-corrected p-values (Liu, Gao et al. Citation2022).
Results
Seven novel SNPs identified
Phenotypic data (the number of thoracolumbar number and carcass traits) of 406 donkeys are shown in Table S2. The total number of thoracic and lumbar vertebrae was 23 in most cases and 87.7% in our population.
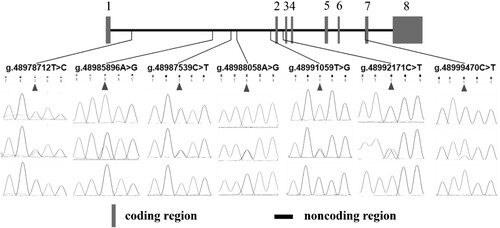
Targeted sequencing results showed that a total of 111 SNPs were identified (Table S3). Among them, 4 SNPs were located in exons, 93 SNPs were located in introns, 5 SNPs were located upstream of DCAF7 genes, 6 SNPs were located downstream of DCAF7 genes and 3 SNPs were located in intergenic regions. However, 105 SNPs had a genotype frequency of less than 5%, therefore statistics was not applied to these data. A total of seven SNPs were analysed (g.48978712 T > C, g.48985896 A > G, g.48987539 C > T, g.48988058 A > G, g.48991059 T > G, g.48992171 C > T and g.48999470 C > T). Among them SNP g.48978712 T > C, g.48985896 A > G, g.48987539 C > T, g.48988058 A > G and g.48991059 T > G were located in the intron 1, g.48992171 C > T was located in the intron 2 and g.48999470 C > T was located in the intron 6 (). The genotyping results of seven SNPs of DCAF7 gene in 406 Dezhou donkeys are shown in Table S4.
Genetic parameter analysis
The allelic frequencies, genotypic frequencies and population genetic parameters (Ho, He, Ne, PIC and HWE) of the seven SNPs in the donkey DCAF7 gene were shown in . Seven SNPs were all dominated by the normal allele. Among the seven SNPs, g.48999470 C > T had the highest normal allele frequency with an allele frequency of 0.7869, and had a highest frequency of normal genotypes, with genotype frequencies of 0.6355(). g.48991059 T > G had the lowest normal frequency with a frequency of 0.5739, and had the lowest frequency of normal genotype with 0.3547 (). Among the seven SNPs, g.48978712 T > C, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T had the same mutant allele frequency with an allele frequency of 0.2204, and had the lowest mutant genotype frequencies, all of which were 0.0567 ().
Table 2. Genetic parameters of seven SNPs in the DCAF7 gene in Dezhou donkey.
In the Hardy–Weinberg equilibrium test, all loci were in the Hardy-Weinberg equilibrium except for g.48991059 T > G (P > 0.05). The differences between Ho and He were higher than 0.3 and lower than 0.4 for all locus except g.48991059 T > G. g.48991059 T > G had the highest He, Ho, Ne and PIC of 0.5085, 0.4915, 1.9664 and 0.3707, respectively (). g.48999470 C > T had the lowest He, Ho, Ne and PIC of 0.6647, 0.3353, 1.5045 and 0.2791, respectively (). Seven sites were in moderate polymorphism (0.25 < PIC < 0.5). This reflects that the genetic diversity of DCAF7 gene in Dezhou donkeys is moderate.
Haplotype construction of DCAF7 gene in Dezhou donkey
The linkage disequilibrium between the seven mutation sites of DCAF7 gene were estimated. The analysis revealed the linkage relationship between the seven SNPs of DCAF7 gene in Dezhou donkey (), and the a-plot is D’ and the b-plot is r2. SNPs (g.48978712 T > C, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T) are inherited together, and it was found that these four SNPs could be used as tag SNP. g.48978712 T > C was selected as tag SNP randomly. Linkage disequilibrium analysis of g.48978712 T > C, g.48985896 A > G, g.48991059 T > G and g.48999470 C > T was performed, and the results showed that g.48978712 T > C and g.48985896 A > G (D′ = 0.96/r2 = 0.96), g.48978712 T > C and g.48999470 C > T (D′ = 0.91/r2 = 0.91) had a strong linkage relationship. In addition, there was a strong linkage between g.48985896 A > G and g.48999470 C > T (D′ = 0.88/r2 = 0.88), while g.48991059 T > G had lower linkage to the other three SNPs (r2 < 0.33).
Figure 2. Linkage (D’, r2) on SNPs of the DCAF7 gene in Dezhou donkey. (The a and b plots are haplotypes constructed from seven SNPs for D’ and r2 using Tag SNP, the c and d plots are haplotypes constructed from four strong linkage SNPs for D’ and r2).
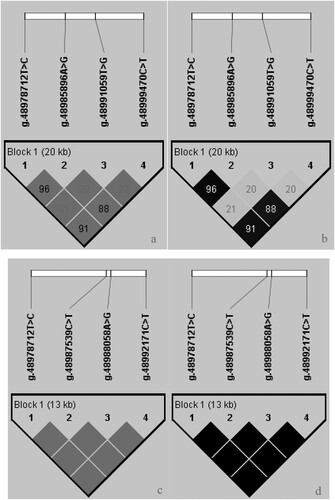
The four SNPs (g.48978712 T > C, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T) of DCAF7 gene were used for haplotype construction: Hap1: TAGC (42.50%), Hap2: TATC (34.70%), Hap3: CGTT (20.70%), Hap4: CGTC (1.10%) (). Hap1 showed the highest haplotype frequencies (42.50%), whereas Hap4 had the lowest haplotypic frequencies (1.10%). Furthermore, the seven haplotype combinations had more than six samples that were involved in the association analysis. Only eight haplotype combinations were found in our population. However, there was only one sample of Hap1Hap4 in our population, so Hap1Hap4 was not used for association analysis.
Table 3. Haplotypes of DCAF7 gene and their frequencies in Dezhou donkey.
Considering that one of the four SNPs (g.48978712 T > C, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T) was randomly able to act as a Tag SNP, indicating that these four SNPs were strong linkage with each other (r2 > 0.33). Therefore, haplotype construction was performed for these four SNPs. The four SNPs of DCAF7 gene were used for haplotype construction: Hap5: TCAC (78.00%), Hap6: CTGT (22.00%) (). In addition, a total of three haplotype combinations were constructed, the three haplotype combinations had more than six samples that were involved in the association analysis.
Table 4. Haplotypes constructed from four SNPs of the Dezhou donkey DCAF7 gene and their frequencies.
Association analysis of DCAF7 SNPs with number of thoracolumbar vertebrae and carcass traits
The effects of the seven SNPs of DCAF7 gene on the number of thoracolumbar vertebrae and carcass traits in Dezhou donkey are summarized in . The different genotypes of g.48978712 T > C, g.48985896 A > G, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T were associated with number of thoracic vertebrae (P < 0.05), and the heterozygote genotype were higher than that for other two genotypes, furthermore, the normal genotype is intermediate between the mutant genotype and the heterozygous genotype. g.48991059 T > G was significantly associated with hide weight, the TT (24.51 ± 2.95 kg) genotype of Dezhou donkey was significantly higher than the genotypes TG (23.76 ± 2.88 kg) and GG (24.40 ± 2.60 kg) (P < 0.05).
Table 5. Association of different genotypes of SNPs in DCAF7 gene with number of thoracolumbar vertebrae and carcass traits in Dezhou donkey.
Association analysis of DCAF7 haplotype combinations with number of thoracolumbar vertebrae and carcass traits
Association analysis of the DCAF7 haplotype combinations with number of thoracolumbar vertebrae and carcass traits in Dezhou donkeys revealed that the length of the thoracic vertebrae differed in different haplotype combinations (P < 0.05) (). The length of thoracic vertebrae in Hap2Hap4 (76.07 ± 2.13 cm) of Dezhou donkey individuals was significantly higher than that in Hap1Hap2 (72.25 ± 3.90 cm) and Hap1Hap3 (72.52 ± 3.52 cm) (P < 0.05).
Table 6. Association of combined haplotypes of seven SNPs with number of thoracolumbar vertebrae and carcass traits in Dezhou donkey.
Association analysis of haplotype combinations constructed from four fully linked SNPs (g.48978712 T > C, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T) of the DCAF7 gene with the number of thoracolumbar vertebrae and carcass traits in Dezhou donkeys showed that the number of the thoracic vertebrae differed in different haplotype combinations (P < 0.05) (). The number of thoracic vertebrae in Hap5Hap6 (17.93 ± 0.33 cm) of Dezhou donkey individuals was significantly higher than that in Hap5Hap5 (17.82 ± 0.40 cm) (P < 0.05).
Table 7. Association of haplotype combinations of four fully linked SNPs with the number of thoracolumbar vertebrae and carcass traits in Dezhou donkeys.
Disscussion
Compared with other livestock, the current development of donkey industry is in its initial stage. Excellent breeds are the basis of modern animal husbandry. However, the current breeding of donkeys is slow, and the development of the donkey industry urgently needs to breed new breeds with meat-rich and hide-thick. Molecular marker technology-assisted breeding can shorten breeding years, speed up the breeding process and improve breeding efficiency.
The SNP of the DCAF7 gene has not been found in donkeys, cows, horses, pigs or sheep. Seven SNPs of the DCAF7 gene were reported for the first time in donkeys, and all of them were located in introns. The eukaryotic intron region has a huge number of SNPs, and SNPs in the first intron are more impactful than those in other introns (Chen et al. Citation2010). In our study, there are five SNPs located in the first intron. SNPs located in introns affect the binding of shearing factors, forming new transcripts that ultimately affect gene expression. Guo et al. (Citation2014) found g. 11043C > T in intron 1 of the SPEF2 gene, which affects the binding of the shear factor binding protein SC35 to the target sequence, which may be responsible for affecting semen traits in bulls. Whether the seven SNPs we identified affect shear factor binding sites needs to be further investigated. The Ho, Ne and PIC values of the seven loci did not differ significantly, suggesting that the degree of variation and selection potential of these loci were essentially the same in the population (Lai et al. Citation2020). The relatively large difference between Ho and He may be due to the presence of inbreeding. The g.48991059 T > G locus is not in the HWE and seven SNPs were in moderate polymorphism, this may be due to artificial selection, or the presence of backcrossing or inbreeding within the population (Gao et al. Citation2019).
Analysis of the association between mutant loci in the DCAF7 gene and livestock production performance has not been studied. In our study, the g.48991059 T > G locus was identified as a candidate locus affecting hide weight, g.48978712 T > C, g.48985896 A > G, g.48987539 C > T, g.48988058 A > G and g.48992171 C > T are the candidate loci affecting the number of thoracic vertebrae. This is consistent with the findings of Bharambe et al. (Citation2020) on the function of the DCAF7 gene, the DCAF7 gene may also play a role in the proliferation and differentiation of vertebrate somatic cells in donkeys by co-activating the Notch signalling system, although more research is needed. The above results indicated that Dezhou donkeys with genotype g.48991059 T > G-TT could be selected to obtain a higher weight of hide and with genotypes g.48978712 T > C-TC, g.48985896 A > G-AG, g.48987539 C > T-CT, g.48988058 A > G-AG and g.48992171 C > T-CT could be selected to obtain number of thoracic vertebrae. In most studies, the most ideal genotype is the normal genotype or mutation genotype, probably because our population is not large enough and probably because our experimental animals come from slaughterhouses with unclear kinship. SNPs located in introns also play an important role in many aspects of economic traits in donkey. DCAF16 gene and DCAF7 gene are the same family and have similar biological functions. Hou et al. (Citation2018) identified a synonymous mutation in exon 2 of the DCAF16 gene, the results of the loci association analysis showed that the body height and chest circumference of individuals with AA and GA genotypes were significantly higher than those of GG genotypes at 6 months of age in Dezhou donkey (P < 0.01). Liu, Gao et al. (Citation2022) found that the NR6A1 gene g.18114954 C > T was associated with the number of lumbar vertebrae and total number of thoracolumbar vertebrae in Dezhou donkeys (P < 0.05). Similarly, our results demonstrated that the DCAF7 gene can be used as a candidate marker for selection for multiple thoracolumbar vertebrae in Dezhou donkeys.
Haplotypes are more useful as genetic markers than SNP locus because they have a higher possibility of being inherited jointly. For those traits with moderate to low heritability, haplotypes were more effective than SNP (Zhao et al. Citation2022). The SNPs we identified were all in the moderate polymorphism. Hap2Hap4 individuals with the longest thoracic vertebrae and can be used as a molecular marker for breeding donkey breeds with longer thoracolumbar vertebrae. Haplotype combinations constructed from introns that are significantly associated with body size traits. It has been extensively reported in other large domestic animals. Huang et al. (Citation2014) discovered haplotype combinations constructed from four SNPs located in the intron that were significantly associated with growth traits in Qinchuan cattle. In the present research, haplotype combinations constructed from seven SNPs were not significantly associated with the number of thoracic vertebrae, but haplotype combinations constructed from four fully linked SNPs (g.48978712 T > C, g.48987539 C > T, g.48988058 A > G, g.48992171 C > T) were associated with the number of thoracic vertebrae, probably influenced by the other three loci (g.48991059 T > G, g.48985896 A > G, g.48999470 C > T). The sample size needs to be further expanded and will require further study at a later stage.
Conclusion
In conclusion, we examined the variation of the DCAF7 gene in the Dezhou donkey population, we demonstrated that the DCAF7 gene in association with number of thoracolumbar vertebrae and carcass traits but the regulatory mechanism of which needs further study. This study verified that the DCAF7 gene could use as a potential molecular marker for donkey genetic breeding and laid the foundation for breeding multi-thoracolumbar vertebrae donkeys.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care Committee at Liaocheng University (Permit No. LC2019-1).
Supplemental Material
Download MS Word (153 KB)Acknowledgments
The authors gratefully acknowledge all participants for their help in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data are available by sending an email to the corresponding author.
Additional information
Funding
References
- Alvarado E, Yousefelahiyeh M, Alvarado G, Shang R, Whitman T, Martinez A, Yu Y, Pham A, Bhandari A, Wang B, Nissen RM. 2016. Wdr68 mediates dorsal and ventral patterning events for craniofacial development. PLoS One. 11(11):e0166984.
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21(2):263–265.
- Bharambe HS, Joshi A, Yogi K, Kazi S, Shirsat NV. 2020. Restoration of miR-193a expression is tumor-suppressive in MYC amplified Group 3 medulloblastoma. Acta Neuropathol Commun. 8(1):70.
- Bonano M, Martín E, Moreno Ruiz Holgado MM, Silenzi Usandivaras GM, Ruiz De Bigliardo G, Aybar MJ. 2018. Molecular characterization of wdr68 gene in embryonic development of Xenopus laevis. Gene Express Patterns. 30:55–63.
- Chen R, Davydov EV, Sirota M, Butte AJ. 2010. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS One. 5(10):e13574.
- Erdenee S, Akhatayeva Z, Pan C, Cai Y, Xu H, Chen H, Lan X. 2021. An insertion/deletion within the CREB1 gene identified using the RNA-sequencing is associated with sheep body morphometric traits. Gene. 775:145444.
- Fan Y, Xing Y, Zhang Z, Ai H, Ouyang Z, Ouyang J, Yang M, Li P, Chen Y, Gao J, et al. 2013. A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and western pigs. PloS One. 8(4):e62534.
- Gao Y, Huang B, Bai F, Wu F, Zhou Z, Lai Z, Li S, Qu K, Jia Y, Lei C, Dang R. 2019. Two novel SNPs in RET gene are associated with cattle body measurement traits. Animals (Basel). 9(10):836.
- Guo F, Luo G, Ju Z, Wang X, Huang J, Xu Y. 2014. Association of SPEF2 gene splices variant and functional SNP with semen quality traits in Chinese Holstein bulls. J Nanjing Agricult Univer. 37(3):119–125.
- Hou H, Li H, Yang L, Zhang X, Shi T, Wang X, Zhao Z, Zhang L. 2018. Association between NCAPG-DCAF16 region polymorphisms and growth traits in Dezhou Donkeys. Acta Vet Zootech Sinica. 50(2):302–313.
- Huang Y, Zhan Z, Li X, Wu S, Sun Y, Xue J, Lan X, Lei C, Zhang C, Jia Y, Chen H. 2014. SNP and haplotype analysis reveal IGF2 variants associated with growth traits in Chinese Qinchuan cattle. Mol Biol Rep. 41(2):591–598.
- Jin Y, Yang Q, Zhang M, Zhang S, Cai H, Dang R, Lei C, Chen H, Lan X. 2019. Identification of a novel polymorphism in Bovine lncRNA ADNCR gene and its association with growth traits. Anim Biotechnol. 30(2):159–165.
- Lai Z, Li S, Wu F, Zhou Z, Gao Y, Yu J, Lei C, Dang R. 2020. Genotypes and haplotype combination of ACSL3 gene sequence variants is associated with growth traits in Dezhou donkey. Gene. 743:144600.
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, et al. 2016. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. 25(13):2862–2872.
- Li C, Zhang X, Cao Y, Wei J, You S, Jiang Y, Cai K, Wumaier W, Guo D, Qi J. 2017. Multi-vertebrae variation potentially contribute to carcass length and weight of Kazakh sheep. Small Rumin Res. 150:8–10.
- Li M, Zhu M, Chai W, Wang Y, Fan D, Lv M, Jiang X, Liu Y, Wei Q, Wang C. 2021. Determination of lipid profiles of Dezhou donkey meat using an LC-MS-based lipidomics method. J Food Sci. 86(10):4511–4521.
- Li Y, Ma Q, Zhou M, Zhang Z, Han Y, Liu G, Zhu M, Wang C. 2022. A metabolomics comparison in milk from two Dezhou donkey strains. Eur Food Res Technol. 248:1267–1275.
- Liu J, Sun S, Han L, Li X, Sun Z. 2010. Association between single nucleotide polymorphism of ActRIIB gene and vertebra number variation in small tail Han sheep. Acta Vet Zootech Sinica. 41(8):951–954.
- Liu Z, Gao Q, Wang T, Chai W, Zhan Y, Akhtar F, Zhang Z, Li Y, Shi X, Wang C. 2022. Multi-thoracolumbar variations and NR6A1 gene polymorphisms potentially associated with body size and carcass traits of Dezhou Donkey. Animals. 12(11):1349.
- Morriss GR, Jaramillo CT, Mikolajczak CM, Duong S, Jaramillo MS, Cripps RM. 2013. The drosophila wings apart gene anchors a novel, evolutionarily conserved pathway of neuromuscular development. Genetics. 195(3):927–940.
- Napolitano G, Malta CD, Esposito A, Araujo MEGD, Pece S, Bertalot G, Matarese M, Benedetti V, Zampelli A, Stasyk T, et al. 2020. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dube syndrome. Nature. 585(7826):597–602.
- Nissen RM, Amsterdam A, Hopkins N. 2006. A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev Biol. 6:28.
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. 2008. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8(3):224–236.
- Sun J, Gao Y, Liu D, Ma W, Xue J, Zhang C, Lan X, Lei C, Chen H. 2012. Haplotype combination of the bovine INSIG1 gene sequence variants and association with growth traits in Nanyang cattle. Genome. 55(6):429–436.
- Wang B, Doan D, Roman PY, Alvarado E, Alvarado G, Bhandari A, Mohanty A, Mohanty S, Nissen RM. 2013. Wdr68 requires nuclear access for craniofacial development. PLoS One. 8(1):e54363.
- Wang C, Li H, Guo Y, Huang J, Sun Y, Min J, Wang J, Fang X, Zhao Z, Wang S, et al. 2020. Donkey genomes provide new insights into domestication and selection for coat color. Nat Commun. 11(1):6014.
- Yang S, He H, Zhang Z, Niu H, Chen F, Wen Y, Xu J, Dang R, Lan X, Lei C, et al. 2020. Determination of genetic effects of SERPINA3 on important growth traits in beef cattle. Anim Biotechnol. 31(2):164–173.
- Yu D, Cattoglio C, Xue Y, Zhou Q. 2019. A complex between DYRK1A and DCAF7 phosphorylates the C-terminal domain of RNA polymerase II to promote myogenesis. Nucleic Acids Res. 47(9):4462–4475.
- Yue J, Guo H, Zhou W, Liu X, Wang L, Gao H, Hou X, Zhang Y, Yan H, Wei X, et al. 2018. Polymirphism sites of TGFβ3 gene and its association analysis with vertebral number of porcine. China Anim Husbandry Vet Med. 45(3):738–744.
- Zhang Z, Zhan Y, Han Y, Liu Z, Wang Y, Wang C. 2021. Estimation of liveweight from body measurements through best fitted regression model in Dezhou donkey breed. J Equine Vet Sci. 101:103457.
- Zhao J, Sauvage C, Bitton F, Causse M. 2022. Multiple haplotype-based analyses provide genetic and evolutionary insights into tomato fruit weight and composition. Hortic Res. 9:uhab009.
- Zheng L, Zhang G, Dong Y, Wen Y, Dong D, Lei C, Qi X, Chen H, Huo L, Huang Y. 2019. Genetic variant of MYLK4 gene and its association with growth traits in Chinese cattle. Anim Biotechnol. 30(1):30–35.