ABSTRACT
Purpose: Human adenoviruses (HAdV) from species A, B and C are commonly recognized as pathogens causing severe morbidity and mortality in hematopoietic stem cell transplant (HSCT) recipients. The purpose of the present study was to determine HAdV types responsible for viremia in HSCT recipients at a large tertiary hospital in Poland.
Methods: Analysis of partial nucleotide sequences of HAdV hexon gene was used to type 40 clinical isolates of HAdV obtained from 40 HSCT recipients.
Results: We identified six different HAdV serotypes belonging to species B, C and E. We demonstrated high variability in sequences of detected HAdV types, and patients infected with the same HAdV types were not hospitalized at the same time, which suggests the low possibility of cross-infection. In almost all patients, anti-HAdV antibodies in IgG class were detected, which indicates a history of HAdV infection in the past. Clinical symptoms accompanying HAdV viremia were in 89%, and in 61.5% of individuals, HAdV was a sole pathogen detected. There were no cases with high-level HAdV viremia and severe systemic or organ infections. Graft-versus-host disease (GvHD) was present in patients infected with species B and C, but grade II of GvHD was observed only in patients infected with HAdV-B.
Conclusions: The predominance of HAdV-C and common presence of anti-HAdV antibodies in IgG class may strongly suggest that most infections in the present study were reactivations of HAdV persisting into the patient’s mucosa-associated lymphoid tissues. Variability of HAdV sequences suggests that cross-infections between patients were very rare.
ABBREVIATIONS: GvHD: graft-versus-host disease; HAdV: human adenoviruses; HSCT: hematopoietic stem cell transplantation
Introduction
Until now, 67 genotypes of human adenoviruses (HAdV) are known, which are classified into 7 species (A–G), and classification is based mainly on genome organization and DNA sequence similarity [Citation1]. Infections with adenoviruses are ubiquitous, and in the immunocompetent population, they usually demonstrate low pathogenicity. On the other hand, HAdV are the pathogens causing severe infections in immunocompromised patients, particularly in allogeneic hematopoietic stem cell transplant (HSCT) recipients. Infection may occur due to primary infection or the activation of endogenous adenovirus persisting in several types of cells [Citation2]. In patients subjected to HSCT, HAdV infection is commonly associated with respiratory tract infections, gastroenteritis or hemorrhagic cystitis. Respiratory tract infections are associated with species A, B, C and E, and range from common cold or acute pharyngitis to severe pneumonitis. Gastrointestinal infections, typically presenting with diarrhea, are caused mainly by HAdV from species F and G. Hemorrhagic cystitis is frequently associated with HAdV-B [Citation3]. HAdVs from species A, B and C are commonly recognized as pathogens causing severe morbidity and mortality in immunocompromised patients [Citation4,Citation5].
The aim of the present study was to determine HAdV types responsible for infections in HSCT recipients hospitalized in the largest clinical setting in Poland.
Materials and methods
The material comprised 40 sera samples, determined previously as positive for HAdV DNA with real-time PCR, taken from 40 adult patients, who underwent allogeneic hematopoietic stem cell transplantation in the Department of Haematology, Oncology and Internal Medicine, Medical University of Warsaw between 2007 and 2012 [Citation6]. Patients’ characteristics are shown in .
Table 1. Basic characteristics of patients from study group (N = 40).
The presence of IgG antibodies specific for HAdV was measured using a commercial NovaLisa™ ELISA qualitative test (NovaTec Immundiagnostica, Dietzenbach, Germany) according to the manufacturer's instructions.
Total DNA was extracted from 200 μl of serum using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Germany), following the manufacturer's instructions. Real-time PCR tests used for the detection of adenoviral DNA in sera samples were run on the LightCycler 2.0 instrument (Roche Diagnostics, Mannheim, Germany) using a modified in-house quantitative method, which targets the hexon gene and allows detection of all known species of HAdV [Citation6].
For sequencing, the methodology developed by Sarantis et al. [Citation7] was used, including reaction parameters and sequences of primers, covering conserved region and bracketing HVR-7 (hypervariable region) of the HAdV hexon gene. PCR was run on the GeneAmp® PCR 9700 instrument (Applied Biosystems, Foster City, CA, U.S.A.) with subsequent electrophoresis of reaction products in 1.5% agarose gel with visualization in an ultraviolet transilluminator. Later, amplicons were purified with Exo-SAP (Invitrogen, Carlsbad, CA, U.S.A.), denatured and sequenced with sense and antisense primers using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing analysis was carried out using ABI 3130 capillary analyzer (Applied Biosystems). For further analysis, 586 bp upstream and downstream consensus sequences of the amplified region were used.
Obtained sequences were compared to the NCBI database using the BLAST software. Homologous highly similar sequences of previously published HAdVs with the highest scores were considered as identification results of HAdV type and species. For multiple comparisons of the sequences, the DAMBE version 5.0.25 (Xuhua Xia, University of Ottawa, Canada) was used [Citation8].
Statistical analysis was performed using the Statistica 10 software (StatSoft, Inc., Krakow, Poland). χ2 test was used to examine the populational associations. p-Values <0.05 were considered to be statistically significant.
Results
Among 40 sequenced HAdV isolates, we identified 6 different types belonging to 3 species. The most frequently detected species was HAdV-C (57.5%), followed by species B (35%) and E (7.5%). Other adenovirus species were not detected. We did not find co-infection with different types of HAdV in a described group of patients.
From species C, HAdV-2 was detected in 15 patients (37.5%), HAdV-5 in 8 patients (15%) and HAdV-6 in 2 patients (5%). From species B, HAdV-3 was detected in 8 patients (20%) and HAdV-7 in 6 patients (15%). HAdV-4 was identified in 8 patients (7.5%). Possibility of HAdV cross-infection was on very low level, because patients infected with the same HAdV types were not hospitalized at the same time (, ). Interestingly, in 39 patients (97.5%), anti-HAdV antibodies in IgG class were detected, which indicates a history of HAdV infection in the past. Most HAdV infections could be therefore reinfections or reactivations of persistent virus. HAdV viral loads, measured in blood, ranged from 1.03 × 103 to 2.86 × 104 copies/ml (median 1.83 × 103 copies/ml, ). HAdV viremia was detected both in early and late post-transplantation periods, starting as early as the 2nd day after transplantation, up to the 209th day after transplantation; the median was 46.5 days.
Figure 1. Nucleotide sequence-based comparison of clinical strains of HAdV. Strains described with sample number and type represent HAdV from this study. Names in frames represent reference sequences from NCBI database, with NCBI accession number and HAdV type indicated.
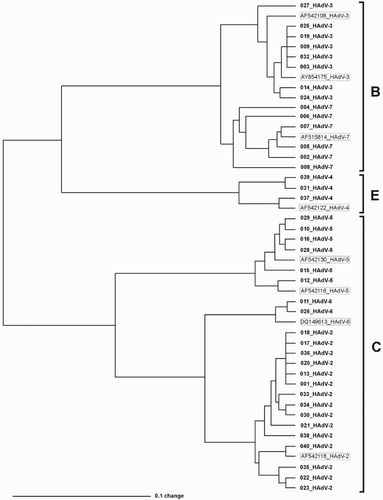
Table 2. Association between HAdV serotype, its viral load and selected clinical parameters.
Symptoms accompanying HAdV viremia manifested usually as respiratory tract infections (11 patients, 27.5%) with cough as the most common symptom (20%). Thirty-six of patients (90%) developed fever.
In over one-third of patients, HAdV viremia was accompanied by infections with other pathogens. Bacterial infections were detected in 7 patients (17.5%), fungal infections in 2 patients (5%) and cytomegalovirus (CMV) viremia was identified in 5 patients (12.5%). Mixed infections of multiple etiology were diagnosed in four patients (). In the remaining 21 patients (52.5%), HAdV was the sole pathogen detected at the time.
Cough was the most common symptom (20%) of respiratory tract infection (RTI) and was observed mostly in patients infected with HAdV-2 and HAdV-3. More severe localized symptoms were observed mostly in patients with other coexisting infections: two patients, both infected with HAdV-2, had pneumonia of bacterial origin; two patients infected with HAdV-3, and HAdV-2 had bronchitis – in the latter case accompanied with C. albicans and CMV systemic infection. Diarrhea was observed in one patient, infected with HAdV-2, but it was most probably caused by C. difficile, detected in stool at the same time. Four patients (10%) had viremia without any clinical manifestations and seven patients were lymphopenic. Eight patients (20%) developed low-grade GvHD – all cases presented with skin manifestations (erythematous rash); five of them were infected with HAdV-B, while others with HAdV-C. Patients infected with HAdV-B showed GvHD grade I and II, and patients infected with HAdV-C showed only GvHD I°. However, there was no statistically significant association between HAdV species and GvHD (p = 0.137).
Discussion
In this study, 40 clinical isolates were typed using the sequencing assay. According to other authors, species C is the most common cause of HAdV infection in HSCT recipients [Citation9–11]. Similarly, most of the infections in our study (57.5%) were caused by HAdV-C. HAdV-C are known to establish an asymptomatic persistent infection in immunocompetent individuals, particularly in children [Citation12. After primary infection, HAdV-C may persist in mucosa-associated lymphoid tissue and viral shedding may occur during immunosuppression. Garnett et al. found that mainly serotypes 1, 2 and 5 persist in tonsils for years [Citation13]. In our study, HAdV-2 was the most frequently detected serotype (37.5%). We also detected the most common members of species B, HAdV-3 and HAdV-7 in 20 and 15% of patients, respectively. GvHD was present in patients infected with species B and C, but grade II of GvHD was observed only in patients infected with HAdV-B. Lion et al. showed that most patients who presented GvHD were infected with species C, but they also have not found an association between HAdV species and GvHD [Citation5].
It is well known that not all HAdV-infected immunocompromised patients develop clinical symptoms [Citation14]. We found that 10% of individuals with HAdV DNA viremia level >1000 c/ml have not developed any clinical manifestations. The most common clinical symptom was fever, present in 90% of patients in our study group and in almost two-thirds of patients it was the only symptom of infection, but it is known that fever may be the only sign of infection in the immunocompromised patients [Citation15]. Moreover, in 11 patients (27.5%) other factors responsible for fever development have been observed: CMV viremia, bacterial urinary tract infection, leukopenia and bacteremia. Concerning localization of clinical symptoms, respiratory tract infections were the most apparent, but again, pneumonia observed in two patients had bacterial background, whereas bronchitis and cough, present in other two patients, had confirmed mixed bacterial–fungal origin in one case, and the second case represented the single lower RTI where no other pathogen was detected at the time of HAdV viremia, which may indicate HAdV-3 as a causative agent.
Mixed infections of adenoviral and bacterial/fungal etiology in HSCT recipients have not been yet specifically analyzed. Only recently, there were reports published, presenting cases of disseminated HAdV infections in HSCT recipients accompanied by systemic/organ fungal, bacterial and CMV infections [Citation16,Citation17]. However, in contrast to the present study, described cases were characterized by high-level adenoviral viremia with multiorgan involvement and poor outcome, while HAdV infections analyzed here presented with the mild clinical course, with low to moderate blood viral load, and without mortality attributed to HAdV infection. Still, interesting is that the most common types of adenoviruses identified, HAdV-2 and HAdV-3, presented different picture in terms of coinfections. While HAdV-2 was accompanied by other pathogens in 8 out of 14 cases, HAdV-3 was a single pathogen detected in 7 out of 8 patients. While the assumption can be made that described high level of coinfections may result from interactions between HAdV-2 and other pathogens, a small number of observations do not allow for any conclusion at this point.
Adenoviral viremia has been recognized as an independent risk factor of HAdV symptomatic disease, and severity of symptoms has been shown to be positively correlated with blood viral load as measured with quantitative real-time PCR [Citation18]. According to the latest approach to adenoviral infections in HSCT settings, there are two major factors influencing the risk of symptomatic disease: time from transplantation to HAdV viremia onset and initial HAdV viral load [Citation19]. In the presented study group, there were 4 patients with HAdV types 2, 3 and 7 blood viral load over 10,000 copies/ml; 2 of them had infections in early post-transplantation period (HAdV-3 at 27th and HAdV-7 at 28th day), and 2 of them – in late post-transplantation period (62nd and 105th day, both caused by HAdV-2). All four patients developed clinical symptoms at the time of HAdV viremia, and three of them had also other infections – bacteremia accompanied by CMV infection, bacterial pneumonia and invasive fungal infection, respectively. In one patient, HAdV was responsible for febrile illness observed at the time of viremia in the early post-transplantation period. Thus, there is a possibility of both early and late infections with elevated levels of HAdV DNA in blood. Despite higher, but nonsignificant frequency of HAdV infections in early versus late post-transplantation period (28 versus 12 patients, respectively, p = 0.059), there were no significant differences in development of high- and low-copy infection, symptomatic disease, nor the presence of concomitant infections; however, the small size of the patients’ group and small difference in viral load may be also a possible reason. Similarly, there was no correlation between these parameters and both donor source (sibling versus matched unrelated donor) and immunosuppression used (myeloablative versus nonmyeloablative).
It is possible that some persons are being exposed to new HAdV serotypes during their hospitalization, but in the present study, there was no evidence of such cross-infection. Nosocomial outbreak of HAdV-31 infection in a pediatric hematology unit has been reported previously [Citation20], however in our study, no patient was infected with HAdV-A. The predominance of HAdV-C and the common presence of anti-HAdV antibodies in IgG class may strongly suggest that most infections in the present study were reactivations of HAdV persisting in patients’ tissues.
This study has many limitations. Particularly, due to its retrospective nature, there was no possibility to examine urine, stool and respiratory specimens, even though information about viral load in these materials could be very useful, especially in terms of adenovirus involvement in etiology, especially in cases of mixed infections and in terms of coinfections with different types of HAdV as well. Similarly, there was no possibility to monitor the dynamics of HAdV viremia. Finally, a limited number of patients do not allow for more accurate statistical analysis.
The presented molecular epidemiology results showed that adenoviral infections in Polish HSCT setting are caused by HAdV from B, C and E species, but there is the clear genetic difference between typed adenoviral strains, which indicates low probability of cross-infections. We also showed that coinfections with adenoviruses and other pathogens (viral, bacterial and fungal) are common, and the possible role of HAdV in etiopathogenesis of these infections cannot be ruled out. This information may help clinical personnel to introduce measures related to diagnosis and treatment of HAdV infections.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Maciej Przybylski http://orcid.org/0000-0002-0868-0018
Anna Waszczuk-Gajda http://orcid.org/0000-0001-5626-1750
Grzegorz W. Basak http://orcid.org/0000-0003-3858-8180
Additional information
Funding
References
- Matsushima Y, Shimizu H, Kano A, et al. Genome sequence of a novel virus of the species human adenovirus D associated with acute gastroenteritis. Genome Announc. 2013;1:e00068-12. doi: https://doi.org/10.1128/genomeA.00068-12
- Echavarría M. Adenoviruses in immunocompromised hosts. Clin Microbiol Rev. 2008;21:704–715. doi: https://doi.org/10.1128/CMR.00052-07
- Ison MG. Adenovirus infections in transplant recipients. Clin Infect Dis. 2006;43:331–339. doi: https://doi.org/10.1086/505498
- Rynans S, Dzieciątkowski T, Młynarczyk G. Adenovirus infection in immunocompromised patients. Postepy Hig Med Dosw (Online). 2013;67:964–972. Polish. doi: https://doi.org/10.5604/17322693.1066199
- Lion T, Baumgartinger R, Watzinger F, et al. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood. 2003;102(3):1114–1120. doi: https://doi.org/10.1182/blood-2002-07-2152
- Rynans S, Dzieciątkowski T, Przybylski M, et al. Incidence of adenoviral DNAemia in Polish adults undergoing allogeneic haematopoietic stem cell transplantation. Arch Immunol Ther Exp (Warsz). 2015;63(1):79–84. doi: https://doi.org/10.1007/s00005-014-0320-z
- Sarantis H, Johnson G, Brown M, et al. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J Clin Microbiol. 2004;42:3963–3969. DOI: https://doi.org/10.1128/JCM.42.9.3963-3969.2004
- Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 2013;30:1720–1728. doi: https://doi.org/10.1093/molbev/mst064
- Chakrabarti S, Mautner V, Osman H, et al. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood. 2002;100(5):1619–1627. doi: https://doi.org/10.1182/blood-2002-02-0377
- Robin M, Marque-Juillet S, Scieux Cet al. Disseminated adenovirus infections after allogeneic hematopoietic stem cell transplantation: incidence, risk factors and outcome. Haematologica. 2007;92(9):1254–1257. doi: https://doi.org/10.3324/haematol.11279
- Ganzenmueller T, Buchholz S, Harste G, et al. High lethality of human adenovirus disease in adult allogeneic stem cell transplant recipients with high adenoviral blood load. J Clin Virol. 2011;52(1):55–59. doi: https://doi.org/10.1016/j.jcv.2011.06.005
- Mynarek M, Ganzenmueller T, Mueller-Heine A, et al. Patient, virus, and treatment-related risk factors in pediatric adenovirus infection after stem cell transplantation: results of a routine monitoring program. Biol Blood Marrow Transplant. 2014;20(2):250–256. doi: https://doi.org/10.1016/j.bbmt.2013.11.009
- Garnett CT, Erdman D, Xu Wet al. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol. 2002;76:10608–10616. doi: https://doi.org/10.1128/JVI.76.21.10608-10616.2002
- Watson T, MacDonald D, Song X, et al. Risk factors for molecular detection of adenovirus in pediatric hematopoietic stem cell transplantation recipients. Biol Blood Marrow Transplant. 2012;18:1227–1234. doi: https://doi.org/10.1016/j.bbmt.2012.01.013
- Öhrmalm L, Wong M, Aust C, et al. Viral findings in adult hematological patients with neutropenia. PLoS One. 2012;7(5):e36543. doi: https://doi.org/10.1371/journal.pone.0036543
- Hubmann M, Fritsch S, Zoellner AK, et al. Occurrence, risk factors and outcome of adenovirus infection in adult recipients of allogeneic hematopoietic stem cell transplantation. J Clin Virol. 2016;82:33–40. doi: https://doi.org/10.1016/j.jcv.2016.07.002
- Engelmann I, Coiteux V, Heim Aet al. Severe adenovirus pneumonia followed by bacterial septicaemia: relevance of co-infections in allogeneic hematopoietic stem cell transplantation. Infect Disord Drug Targets. 2016;16:69–76 doi: https://doi.org/10.2174/1871526516666160407114623
- Lee YJ, Prockop SE, Papanicolaou GA. Approach to adenovirus infections in the setting of hematopoietic cell transplantation. Curr Opin Infect Dis. 2017;30:377–387. doi: https://doi.org/10.1097/QCO.0000000000000379
- Öhrmalm L, Lindblom A, Omar H, et al. Evaluation of a surveillance strategy for early detection of adenovirus by PCR of peripheral blood in hematopoietic SCT recipients: incidence and outcome. Bone Marrow Transplant. 2011;46(2):267–272. doi: https://doi.org/10.1038/bmt.2010.86
- Leruez-Ville M, Chardin-Ouachée M, Neven B, et al. Description of an adenovirus A31 outbreak in a paediatric haematology unit. Bone Marrow Transplant. 2006;38(1):23–28. doi: https://doi.org/10.1038/sj.bmt.1705389
