ABSTRACT
Objectives: E26 transformation-specific variant 6 gene (ETV6) is one of the most consistently rearranged genes in acute leukaemia. It encodes a principal hematopoietic transcription factor.
Methods: We performed a systematic review focusing on the mechanisms responsible for etv6 acquisition, and its effect on the development of AML. We also review the Characteristics of ETV6 mutations and its fusion genes. Finally, for using ETV6 as a molecular target, we discuss future therapeutic approaches available to mitigate the associated disease.
Results: ETV6 rearrangements often accompany other molecular mutations. Thirty-three distinct partner bands of ETV6 that contain various fusion genes were detected which plays a vital role in obtaining information about leukaemia genesis. RXDX-101 and PKC412 were reported to be inhibitors of ETV6-NTRK3.
Discussion: Future researches are needed to explain how ETV6 mutations act within the microenvironment of leukemic cells and how it affects the progression of leukaemia.
Introduction
Acute myeloid leukaemia (AML) is a well-known hematologic malignancy that has a molecular and clinical heterogeneity. Combining chemotherapy and hematopoietic stem cell transplantation are fundamental treatment strategies. In the past 40 years, almost no progress in standard treatment for AML. Long-term survival rates are 25–70% in patients inferior to 60 years and only 5–15% in older patients. Currently, cytogenetic investigation at the time of diagnosis gives the most critical prognostic information, predicting relapse rate, overall survival(OS) and outcome after induction chemotherapy [Citation1]. The ETV6 gene plays an important role in hematopoiesis and hematological malignancies and has the function of tumor suppressive [Citation2].
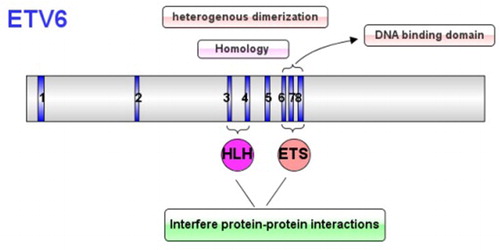
Structure of gene
The ETV6, mapping to chromosome 12p13 in humans, contains 8 exons, is a transcriptional repressor in the ETS transcription factor family. It encodes for a protein with 452 amino acid, containing the unique ETS domain and the HLH domain. The HLH domain is encoded by the third and fourth exons which mediate the homology of ETV6 or the heterogenous dimerization of other ETS family members, and the ETS domain is encoded by the 6th to 8th exons which can specifically combine with the DNA region rich in the purine. Both of the two domains can interfere protein-protein interactions () [Citation2–5]. The ETV6 also contains the N-terminal pointed (PNT) region which induces constitutive activation of tyrosine kinase action [Citation6,Citation7].
Discovery of ETV6 mutations in AML
ETV6 translocates to several partner genes in diverse cancers. Previous studies have informed that ETV6 fusion genes express in mesoblastic nephroma, secretory breast carcinoma, congenital fibrosarcoma and acute leukaemia(ALL and AML) [Citation8–11]. Translocations generate chimeric genes encoding for fusion product (ETV6-ANLN, ETV6-NTRK3, and ETV6-LPNX, e.g.). Previous work indicated that fusion products containing ETV6 involve in numerous regulated errant mechanisms and can influence the fusion partner’s activity through the HLH domain [Citation12]. The ETV6 gene rearrangements were found in a translocation t(12;15)(p13;q25) via Hybridization with cosmid probes which was found in an adult acute myeloid leukaemia (AML) patient with the use of fluorescence in situ hybridization (FISH) method in 1999 [Citation13]. In 2012, Ding L and his union performed whole-genome sequencing of primary tumor and relapse genomes from eight AML patients and certified hundreds of somatic mutations. It is suspected that one or more of the mutations provide a strong particular advantage that contributed to relapse. The ETV6 mutation is one of the most likely candidates they mentioned [Citation14].
Role of ETV6 mutants in leukemogenesis
About 5% of AML and myelodysplastic syndromes have the abnormalities of the short arm of chromosome 12 (12p) which mostly consist of entire or partial loss of 12p usually affecting ETV6, suggesting these genes has a tumor suppressive function [Citation2,Citation15–17]. Likewise, Yamagata et al. mentioned that ETV6 can cause growth restriction and apoptosis [Citation18]. Also, loss of repressor activity was found in AML when heterozygous mutations of ETV6 exist. All these findings add to the view that ETV6 may have tumor-suppressor features.
Alteration of transcription portion function, activation of a proto-oncogene in the section of the translocation, loss of fusion gene function, constitutive kinase activation of the partner protein and the negative effect of the ETV6 fusion protein upon wild-type ETV6-mediated transcriptional suppression are some ways exist in the progress of leukemogenesis when etv6 are involved [Citation11]. Nonreceptor tyrosine kinases, transcription factors, receptor tyrosine kinases and homeobox proteins are some kinds of the products of fusion partner genes [Citation2,Citation11]. Many studies of fusions with protein tyrosine kinases indicated that the PNT filed of ETV6 works as an oligomerization component of the fusion protein, stimulating the tyrosine kinase [Citation6,Citation7]. Constitutive signaling downstream of fused tyrosine kinases plays a significant role in the initiation and maintenance of the malignant phenotype [Citation19].
Previous studies have also suggested that ETV6 mutations usually associated with NPM1 mutations (The role of etv6 together with other genes in the development of leukaemia will be discussed later in this article) [Citation20]. We thus suspect that ETV6 mutations may cooperate with other mutations to establish their own network structure, driving the self-renewal of leukaemia stem cells, thereby promoting the development of leukaemia.
Characteristics of ETV6 mutations
Type, frequency, and stability of ETV6 mutations
ETV6 locates on band 12p13 which are genetically unstable and thus susceptible to chromosomal rearrangements [Citation21]. A study investigated 9550 myeloid disorders patients and identified 0.5% patients with ETV6 rearrangements. Frequencies in AML is 1.1% (40 of 3798). Compared with other subgroups, the FAB M0 subtype, expression of CD7 and CD34, s-AML and t-AML were higher in ETV6 rearranged AML than in de novo AML (). Another molecular analysis suggests that ETV6 rearrangements higher rotation in treatment-related AML [Citation22].
Table 1. Frequency of etv6 rearrangement in patients with different types of AML.
ETV6 fusion gene in AML
Numerous hematological diseases have the fusions between ETV6 and phosphotyrosine kinases (PTKs) and the HLH domain of ETV6 causes abnormal functions [Citation2]. Thirty-three distinct partner bands of ETV6 that contain various fusion genes were detected which plays a vital role in obtaining information about leukaemia genesis. Here we review 3 different types fusion genes.
ETV6 LPXN gene
Akihiro Abe reported ETV6 -LPXN fusion gene in a blood sample of a relapse AML patient whose leukaemia blasts gained t(11;12)(q12.1;p13) at molecule level, but the fusion gene lacked the transcription activation domain [Citation19].
LPXN plays a significant role in cell migration and conglutination [Citation23]. Akihiro Abe pointed that the ETV6-LPXN-transduced 32D and blasts increased migration to the chemokine released by leukaemia cells and the appearance of mRNAs which encode chemokines and conglutination molecules increased at relapse [Citation19,Citation24,Citation25]. On the one hand, ETV6-LPXN may associate with progression of leukaemia through regulating the microenvironment of blasts, on the other hand, it may involve in the relapse of leukaemia.
ETV6-ANLN gene
Campregher P V discovered the etv6-anln fusion gene in the progression of primary myelofibrosis (PMF), which eventually developed into acute myeloid leukaemia (FAB M7).
The ETV6-ANLN fusion gene encodes a protein which almost equals to ANLN and substitutes the first 6 ANLN amino acids by the first 11 ETV6 amino acids, and it loses all ETV6 working fields. Further researches are needed to verify whether this change disturbs the ANLN function and how it works with the missing of the etv6 gene to promote the leukemogenic. Previous studies have demonstrated the function of ANLN in cell separation and DNA synthesis that increased cellular proliferation caused by deregulated ANLN expression may contribute to the development of leukemic malignancies [Citation26,Citation27].
ETV6-NTRK3 gene
The NTRK3 gene, located at chromosome 15q25, includes 18 exons. It encodes for the transmembrane receptor of neurotrophic factor 3 and shows expression in neural tissues as also as in hematopoietic and epithelial cells [Citation28]. The ETV6-NTRK3 fusion gene expresses at low levels in AML and has a strong transforming effect on several cell lines, including hematopoietic cells, and transformed cells can induce tumors in nude mice.
Previous studies reported 2 cases of AML patients, one is AML M2 and the patient associated with severe myelofibrosis at the time of diagnosis, and leukaemia cells quickly transferred to multiple organs, another one was diagnosed with the primary myelofibrosis (PMF) and finally progressed to AML M7.E TV 6 - NTR K3 fusion gene causes cell transformation may be the pathogenesis of acute myeloid leukaemia.
Relationship between ETV6 mutations and other mutations
ETV6 rearrangements often accompany other molecular mutations. The mutation rate of NPM1 and RUNX1 in AML patients with ETV6 rearrangements is respectively 23% and 19%. Genes with a relatively low mutation rate are FLT3-TKD, FLT3-ITD, and MLL-PTD, and the mutation rate is 8.8%, 7.5%, and 4.0% [Citation22]. As additional cytogenetic alterations often appear with ETV6 mutations, further researches are needed to study whether and how do other genes affect ETV6 mutations and how etv6 mutations and other mutations work together to influence the progression of leukaemia.
Therapeutic target and future perspectives
Although several technologies can be used to investigate gene mutations, the efficiency of conventional cytogenetic analysis is restricted because it does not show the status of the partner gene(s) involved in the translocation case. Sequencing by Sanger sequencing to distinguish candidate genes is not only time consuming but costly [Citation29]. Anchored multiplex PCR next-generation sequencing (AMP-NGS) gives the advantage of novel fusion detection and the capability to multiplex multitudinous genes [Citation30]. Applying this technology to patients with AML can help formulating therapeutic strategies.
RXDX-101(also known as Entrectinib) was reported to be an inhibitor of ETV6-NTRK3. TRK kinase inhibition applied to TRK fusion-related hematological malignancies was demonstrated by Kristen M. Smith. Recent preclinical studies demonstrate Entrectinib as a therapy for patients with TRK fusion-involved AML and other hematologic malignancies. It can induce apoptotic cell death and prevent cell proliferation. In models of malignancies, Entrectinib has been shown to regress tumor and destroy residual BM cancer cells [Citation31].
PKC412 is also an inhibitor of ETV6-NTRK3, also known as midostaurin. Hoang ThanhChi demonstrated PKC412 as another inhibitor of ETV6-NTRK3. PKC412 can efficiently inhibit the EN-dependent growth, kinase activity and activation of downstream signaling effectors. Furthermore, similar effects were obtained in inhibition human cell which lines with expression of the ETV6-NTRK3 fusion gene by PKC412.It is suggested that PKC412 might as the therapeutic medicine for treatment of patients with ETV6-NTRK3 fusion. However, more experiments need to be done to examine the effect of PKC412 on the animal model [Citation32].
Future researches should elucidate the mechanisms of etv6 mutations. Whether they interfere essential processes of differentiation and proliferation early in the progress of leukemic alteration and whether they may also relevant in the induction of chromosomal variability finally prompting the development of clones with other molecular mutations. Whether ETV6 mutations act within the microenvironment of leukemic cells and whether it affects the progression of leukaemia should be considered. The last but not least, ETV6 mutations might be of other therapy related in AML and need more suitable patients materials to investigate further.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Fang Zhou http://orcid.org/0000-0001-8401-0457
Additional information
Funding
References
- Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. doi: https://doi.org/10.1016/S0140-6736(06)69780-8
- Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15(3):162–174. Epub 2005/04/14. PubMed PMID: 15826831. doi: https://doi.org/10.1016/j.semcancer.2005.01.008
- Poirel H, Lacronique V, Mauchauffe M, et al. Analysis of TEL proteins in human leukemias. Oncogene. 1998;16(22):2895–2903. Epub 1998/07/22. PubMed PMID: 9671410. doi: https://doi.org/10.1038/sj.onc.1201817
- Baens M, Peeters P, Guo C, et al. Genomic organization of TEL: the human ETS-variant gene 6. Genome Res. 1996;6(5):404–413. Epub 1996/05/01. PubMed PMID: 8743990. doi: https://doi.org/10.1101/gr.6.5.404
- Hart SM, Foroni L. Core binding factor genes and human leukemia. Haematologica. 2002;87(12):1307–1323. Epub 2002/12/24. PubMed PMID: 12495904.
- Kuno Y, Abe A, Emi N, et al. Constitutive kinase activation of the TEL-Syk fusion gene in myelodysplastic syndrome with t(9;12)(q22;p12). Blood. 2001;97(4):1050–1055. Epub 2001/02/13. PubMed PMID: 11159536. doi: https://doi.org/10.1182/blood.V97.4.1050
- Carroll M, Tomasson MH, Barker GF, et al. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc Natl Acad Sci USA. 1996;93(25):14845–14850. Epub 1996/12/10. PubMed PMID: 8962143; PubMed Central PMCID: PMCPmc26224. doi: https://doi.org/10.1073/pnas.93.25.14845
- Knezevich SR, Garnett MJ, Pysher TJ, et al. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58(22):5046–5048. Epub 1998/11/21. PubMed PMID: 9823307.
- Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. Epub 2002/11/27. PubMed PMID: 12450792. doi: https://doi.org/10.1016/S1535-6108(02)00180-0
- Knezevich SR, McFadden DE, Tao W, et al. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18(2):184–187. Epub 1998/02/14. PubMed PMID: 9462753. doi: https://doi.org/10.1038/ng0298-184
- De Braekeleer E, Douet-Guilbert N, Morel F, et al. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36(8):945–961. Epub 2012/05/15. PubMed PMID: 22578774. doi: https://doi.org/10.1016/j.leukres.2012.04.010
- Wlodarska I, La Starza R, Baens M, et al. Fluorescence in situ hybridization characterization of new translocations involving TEL (ETV6) in a wide spectrum of hematologic malignancies. Blood. 1998;91(4):1399–1406. Epub 1998/03/07. PubMed PMID: 9454771.
- Eguchi M, Eguchi-Ishimae M, Tojo A, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood. 1999;93(4):1355–1363. Epub 1999/02/09. PubMed PMID: 9949179.
- Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. Epub 2012/01/13. PubMed PMID: 22237025; PubMed Central PMCID: PMCPmc3267864. doi:10.1038/nature10738.
- Baens M, Wlodarska I, Corveleyn A, et al. A physical, transcript, and deletion map of chromosome region 12p12.3 flanked by ETV6 and CDKN1B: hypermethylation of the LRP6 CpG island in two leukemia patients with hemizygous del(12p). Genomics. 1999;56(1):40–50. Epub 1999/02/26. PubMed PMID: 10036184. doi: https://doi.org/10.1006/geno.1998.5685
- La Starza R, Stella M, Testoni N, et al. Characterization of 12p molecular events outside ETV6 in complex karyotypes of acute myeloid malignancies. Br J Haematol. 1999;107(2):340–346. Epub 1999/12/03. PubMed PMID: 10583222. doi: https://doi.org/10.1046/j.1365-2141.1999.01724.x
- Sato Y, Suto Y, Pietenpol J, et al. TEL and KIP1 define the smallest region of deletions on 12p13 in hematopoietic malignancies. Blood. 1995;86(4):1525–1533. Epub 1995/08/15. PubMed PMID: 7632960.
- Yamagata T, Maki K, Waga K, et al. TEL/ETV6 induces apoptosis in 32D cells through p53-dependent pathways. Biochem Biophys Res Commun. 2006;347(2):517–526. Epub 2006/07/11. PubMed PMID: 16828711. doi: https://doi.org/10.1016/j.bbrc.2006.06.127
- Abe A, Yamamoto Y, Iba S, et al. ETV6-LPXN fusion transcript generated by t(11;12)(q12.1;p13) in a patient with relapsing acute myeloid leukemia with NUP98-HOXA9. Genes Chromosomes Cancer. 2016;55(3):242–250. Epub 2015/11/07. PubMed PMID: 26542893. doi: https://doi.org/10.1002/gcc.22327
- Unnikrishnan A, Guan YF, Huang Y, et al. A quantitative proteomics approach identifies ETV6 and IKZF1 as new regulators of an ERG-driven transcriptional network. Nucleic Acids Res. 2016;44(22):10644–10661. Epub 2016/09/09. PubMed PMID: 27604872; PubMed Central PMCID: PMCPmc5159545. doi: https://doi.org/10.1093/nar/gkw804
- Manola KN, Georgakakos VN, Margaritis D, et al. Disruption of the ETV6 gene as a consequence of a rare translocation (12;12)(p13;q13) in treatment-induced acute myeloid leukemia after breast cancer. Cancer Genet Cytogenet. 2008;180(1):37–42. Epub 2007/12/11. PubMed PMID: 18068531. doi: https://doi.org/10.1016/j.cancergencyto.2007.09.004
- Haferlach C, Bacher U, Schnittger S, et al. ETV6 rearrangements are recurrent in myeloid malignancies and are frequently associated with other genetic events. Genes Chromosomes Cancer. 2012;51(4):328–337. Epub 2011/12/14. PubMed PMID: 22162288. doi: 10.1002/gcc.21918
- Sahu SN, Nunez S, Bai G, et al. Interaction of Pyk2 and PTP-PEST with leupaxin in prostate cancer cells. Am J Physiol Cell Physiol. 2007;292(6):C2288–C2296. Epub 2007/03/03. PubMed PMID: 17329398. doi: https://doi.org/10.1152/ajpcell.00503.2006
- Bruserud O, Ryningen A, Olsnes AM, et al. Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica. 2007;92(3):332–341. Epub 2007/03/07. PubMed PMID: 17339182. doi: https://doi.org/10.3324/haematol.10148
- Peled A, Tavor S. Role of CXCR4 in the pathogenesis of acute myeloid leukemia. Theranostics. 2013;3(1):34–39. Epub 2013/02/06. PubMed PMID: 23382784; PubMed Central PMCID: PMCPmc3563079. doi: https://doi.org/10.7150/thno.5150
- Suzuki C, Daigo Y, Ishikawa N, et al. ANLN plays a critical role in human lung carcinogenesis through the activation of RHOA and by involvement in the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 2005;65(24):11314–11325. Epub 2005/12/17. PubMed PMID: 16357138. doi: https://doi.org/10.1158/0008-5472.can-05-1507
- Campregher PV, Pereira WO, Lisboa B, et al. Identification of ANLN as ETV6 partner gene in recurrent t(7;12)(p15;p13): a possible role of deregulated ANLN expression in leukemogenesis. Mol Cancer. 2015;14:197. Epub 2015/11/21. PubMed PMID: 26584717; PubMed Central PMCID: PMCPmc4653877. doi: https://doi.org/10.1186/s12943-015-0471-5
- Hisaoka M, Sheng WQ, Tanaka A, et al. Gene expression of TrkC (NTRK3) in human soft tissue tumours. J Pathol. 2002;197(5):661–667. Epub 2002/09/05. PubMed PMID: 12210087. doi: https://doi.org/10.1002/path.1138
- Grossmann V, Tiacci E, Holmes AB, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118(23):6153–6163. Epub 2011/10/21. PubMed PMID: 22012066. doi: https://doi.org/10.1182/blood-2011-07-365320
- Badar T, Johnson L, Trifilo K, et al. Detection of novel t(12;17)(p12;p13) in relapsed refractory acute myeloid leukemia by anchored multiplex PCR(AMP)-based next-generation sequencing. Appl Immunohistochem Mol Morphol. 2017. Epub 2017/02/12. PubMed PMID: 28187034. doi: https://doi.org/10.1097/pai.0000000000000477
- Smith KM, Fagan PC, Pomari E, et al. Antitumor activity of entrectinib, a Pan-TRK, ROS1, and ALK inhibitor, in ETV6-NTRK3-positive acute myeloid leukemia. Mol Cancer Ther. 2018;17(2):455–463. Epub 2017/12/15. PubMed PMID: 29237803. doi: https://doi.org/10.1158/1535-7163.mct-17-0419
- Chi HT, Ly BT, Kano Y, et al. ETV6-NTRK3 as a therapeutic target of small molecule inhibitor PKC412. Biochem Biophys Res Commun. 2012;429(1–2):87–92. Epub 2012/11/08. PubMed PMID: 23131561. doi: https://doi.org/10.1016/j.bbrc.2012.10.087