Abstract
Transforming growth factor β (TGF-β) family members play an extensive role in cellular communication that orchestrates both early development and adult tissue homeostasis. Aberrant TGF-β family signaling is associated with a pathological outcome in numerous diseases, and in-depth understanding of molecular and cellular processes could result in therapeutic benefit for patients. Canonical TGF-β signaling is mediated by receptor-regulated SMADs (R-SMADs), a single co-mediator SMAD (Co-SMAD), and inhibitory SMADs (I-SMADs). SMAD7, one of the I-SMADs, is an essential negative regulator of the pleiotropic TGF-β and bone morphogenetic protein (BMP) signaling pathways. In a negative feedback loop, SMAD7 inhibits TGF-β signaling by providing competition for TGF-β type-1 receptor (TβRI), blocking phosphorylation and activation of SMAD2. Moreover, SMAD7 recruits E3 ubiquitin SMURF ligases to the type I receptor to promote ubiquitin-mediated proteasomal degradation. In addition to its role in TGF-β and BMP signaling, SMAD7 is regulated by and implicated in a variety of other signaling pathways and functions as a mediator of crosstalk. This review is focused on SMAD7, its function in TGF-β and BMP signaling, and its role as a downstream integrator and crosstalk mediator. This crucial signaling molecule is tightly regulated by various mechanisms. We provide an overview of the ways by which SMAD7 is regulated, including noncoding RNAs (ncRNAs) and post-translational modifications (PTMs). Finally, we discuss its role in diseases, such as cancer, fibrosis, and inflammatory bowel disease (IBD).
Keywords:
Introduction
The transforming growth factor beta (TGF-β) family is comprised of over 30 secreted multifunctional polypeptides, including TGF-βs, activins, bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and Müllerian-inhibiting substance (MIS). Members of the TGF-β family are ubiquitously expressed in metazoans and play an extensive role in cellular communication that orchestrates both early development and adult tissue homeostasis (Massagué Citation1998). These proteins are involved in a range of cellular processes, including proliferation, cell death, adhesion, migration and differentiation. Additionally, the biological effects of TGF-β family members are highly cell and context dependent. Aberrant TGF-β family signaling is often associated with a pathological outcome, highlighting the importance of this pathway in maintaining tissue homeostasis. Studies have linked inappropriate functional activation or inhibition of TGF-β family signaling components with a broad spectrum of human diseases, including cancer, fibrosis, developmental-, musculoskeletal-, cardiovascular-, and autoimmune disorders (Akhurst and Hata Citation2012; Morikawa et al. Citation2016; Batlle and Massagué Citation2019; Yu and Feng Citation2019).
TGF-β family member ligands are initially synthesized as latent precursor proteins with an N-terminal propeptide region. After proteolytic processing, the mature, biologically active ligand is obtained (Derynck and Budi Citation2019). TGF-β family members signal via heteromeric complexes of single-pass transmembrane type I, also termed activin-receptor-like kinases (ALKs), and type-II serine/threonine kinase receptors (Heldin and Moustakas Citation2016). Seven type-I receptors have been described in vertebrates: ALK1, ALK2 (also termed ActRI), ALK3 (also termed BMPRIA), ALK4 (also termed ActRIB), ALK5 (also termed TβRI), ALK6 (also termed BMPRIB), and ALK7. Type-I receptors for activin and TGF-β are ALK4 and ALK5, respectively. BMPs signal through ALK1, ALK2, ALK3 and ALK6. Five type-II receptors have been identified: TGF-β type-II receptor (TβRII), activin type II and type IIB (ActRII, and ActRIIB), BMP type-II receptor (BMPRII) and AMH type-II receptor (AMHRII). The type-I and type-II receptors have cysteine-rich extracellular domains through which signaling can be initiated by association with TGF-β family member ligands.
Upon ligand binding, the type-I and type-II receptor intracellular domains associate in a heteromeric complex (Wrana et al. Citation1994). Type-I receptors contain a glycine-serine repeat motif (GS domain), of which certain serine and threonine residues can become phosphorylated by the constitutively active type-II receptor kinase. Subsequently, an intracellular cascade by means of phosphorylation of downstream SMAD effector proteins is triggered. These SMADs can act as transcription factors and are the central mediators in the canonical TGF-β signaling pathway (Derynck et al. Citation1998). The eight vertebrate SMADs are divided into three distinct SMAD subtypes: the receptor-regulated SMADs (R-SMADs, SMAD1, 2, 3, 5, and 8), the co-mediator SMAD (Co-SMAD, SMAD4) and inhibitory SMADs (I-SMADs, SMAD6 and 7). Upon receptor activation, specific R-SMADs are phosphorylated at two carboxy (C)-terminal serine residues by the type-I receptor intracellular kinase activity and undergo formation of heteromeric complexes with SMAD4 (Shi and Massagué Citation2003). SMAD2/3 mediate TGF-β family signaling, while SMAD1/5/8 respond primarily to the BMP family. The activated SMAD complexes then translocate into the nucleus where they activate or repress the transcription of target genes. With the exception of a common spliced isoform of SMAD2, the above-mentioned R- and Co-SMADs are able to directly interact with DNA (Massagué Citation2012). The MAD homology (MH)1 domain of these SMADs facilitates their sequence-specific DNA-binding activity. However, SMADs have limited affinity and specificity for DNA and therefore rely on cooperation with other DNA-binding proteins, such as DNA-binding transcription factors and transcriptional coregulators (Hill Citation2016).
I-SMADs were discovered due to their similarity to previously identified R- and Co-SMADs as they also contain a large conserved MH2 domain (Topper et al. Citation1997). The I-SMADs, SMAD6 and SMAD7, are potent modulators of the TGF-β family. They inhibit TGF-β and BMP signaling in a negative feedback circuit by blocking type I receptor activity through physical interaction. Moreover, SMAD7 is able to target receptors for proteasomal degradation, preventing R- and Co-SMAD complex formation (Miyazawa and Miyazono Citation2017). SMAD6 principally regulates BMP signaling, while SMAD7 is able to repress signaling in both the TGF-β and BMP signaling pathway (Itoh and ten Dijke Citation2007) (Figure 1).
While R- and Co-SMADs are critical in propagating TGF-β family signals from cell surface complexes to the nucleus, inhibition of signaling activity by I-SMADs is essential for maintaining a proper physiological response to TGF-β or BMP signaling. This review focuses on our current understanding of the complex and multifaceted role of SMAD7, as a key player in SMAD signaling inhibition, and as downstream integrator providing crosstalk between the TGF-β family and other signaling pathways. We also highlight recent discoveries focusing on SMAD7 regulation. Particularly, we address the increasing number of publications that demonstrate noncoding RNA-governed regulation of SMAD7. Moreover, we describe its function as revealed by genetic studies in mice and human. And finally, we discuss the role of SMAD7 in disease onset and progression, and possible avenues for therapeutic intervention by targeting SMAD7.
SMAD7 structure and function in TGF-β signaling
Human SMAD7 consists of 426 amino acid residues and is comprised of two globular domains, connected by a relatively short linker region (). SMAD7 interacts with the activated TβRI through its MH2 domain (Hayashi et al. Citation1997; Nakao et al. Citation1997). However, SMAD7 lacks a C-terminal SSXS motif, which is required for phosphorylation of SMAD2 and −3 by the TβRI kinase, (Abdollah et al. Citation1997; Souchelnytskyi et al. Citation1997) suggesting that it is not a substrate for TβRI phosphorylation. SMAD7 inhibits signaling by providing competition for receptor binding, blocking phosphorylation and activation of downstream SMAD2. Thereby, it prevents SMAD2/4 complex formation and translocation of SMAD2 to the nucleus (Hayashi et al. Citation1997). Moreover, SMAD7 recruits E3 ubiquitin ligases such as NEDD4L, SMURF1 and SMURF2 to TGF-β receptors, thereby promoting their ubiquitin-mediated degradation (Kavsak et al. Citation2000; Ebisawa et al. Citation2001; Kuratomi et al. Citation2005). Other mechanisms by which SMAD7 inhibits TGF-β signaling include recruitment of phosphatase GADD34-PP1c to the TβRI and hindrance of SMAD-DNA complex formation (Shi et al. Citation2004; Zhang et al. Citation2007).
Figure 1. SMAD7 negatively regulates TGF-β and BMP signaling through physical interaction with the type I receptor, preventing R- and Co-SMAD complex formation and promoting proteasomal degradation of receptors.
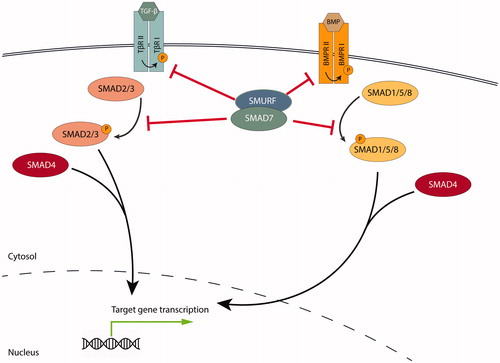
Figure 2. Schematic representation of the structural domains of SMAD7 as they relate to function. PY, proline-tyrosine motif; MH2, Mad homology 2 domain; L3/β8, L3/β8 loop-strand module.
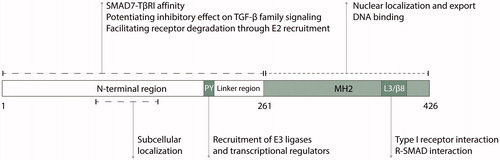
The MH2 domain of SMAD7 contains a basic region that includes the so-called L3 loop and β8 strand (Yan et al. Citation2016). Additionally, unlike R- and Co-SMADs, the SMAD7 MH2 domain also includes an α helix containing K312 and K316, which contributes to the basic character of this domain. SMAD7-TβRI interaction relies on four basic amino acid residues in the basic surface of this C-terminal region: K312, K316, K401, and R409 (Mochizuki et al. Citation2004). Mutations of these residues abrogate the inhibitory effects of SMAD7 on TGF-β signaling by interrupting SMAD7–TβRI interaction. Furthermore, inhibition of BMP signaling by SMAD7 is likely dependent on two of the above-mentioned SMAD7 residues (K401 and R409), as mutation in either residue abolishes SMAD7-mediated inhibition of the pathway (Mochizuki et al. Citation2004).
In addition to facilitating SMAD-receptor interaction, the MH2 domain also enables SMAD-protein interaction (Massagué and Wotton Citation2000). The MH2 domain allows for SMADs to interact with each other and with regulator and effector proteins, including DNA-binding cofactors and chromatin modifiers. The L3/β8 loop-strand module and adjacent lysines comprise key residues that permit SMAD7-SMAD3 association (Yan et al. Citation2016). This suggests that the MH2 domain is not only a critical structural determinant in establishing SMAD-receptor interaction, but also in establishing SMAD-SMAD interaction. Molecular dynamics (MD) simulations predicted that the L3/β8 loop contributes to the considerable functional flexibility of SMAD7. Hariharan et al. describe that SMAD7 has a flexible overall folding, due to the presence of flexible amino acids in the MH2 domain of the protein, and more specifically in the L3 loop (F411, K401, and C406) (Hariharan and Pillai Citation2008).
Additionally, the MH2 domain of SMAD7 is able to bind to oligonucleotides containing the minimal SMAD binding element (SBE). Shi et al. demonstrated that the isolated MH2 domain of SMAD7 had a higher DNA binding affinity than full-length SMAD7 (Shi et al. Citation2008). The difference in binding affinity was attributed to an repressive effect of the SMAD7 N-terminal region. The MH2 domain of SMAD7 is able to bind to DNA and thereby suppresses SMAD2/4-DNA complex formation and is able to directly bind to the plasminogen activator inhibitor 1 (PAI-1) promoter under physiological conditions (Zhang et al. Citation2007).
As described above, SMAD7 inhibits TGF-β and BMP signaling by promoting receptor turnover through ubiquitination (Kavsak et al. Citation2000; Ebisawa et al. Citation2001; Murakami et al. Citation2003; Wiesner et al. Citation2007). The PPxY sequence (proline-tyrosine motif, PY motif) in the SMAD7 linker region has a pivotal role in the recruitment of Homologous to E6AP C-terminus (HECT) type E3 ubiquitin ligases, including NEDD4L, and SMURF1/2 () (Aragón et al. Citation2012). The WW domains of these E3 ligases bind to the PY motif in the linker region of SMAD7. Consistent with this notion, a PY motif deletion or mutation weakens association of SMAD7 with SMURF1/2 (Kavsak et al. Citation2000; Ebisawa et al. Citation2001). It has been reported that the SMURF2 WW3 domain binds to an extended binding motif in SMAD7 that includes the PY motif and six C-terminal residues (Chong et al. Citation2006). Furthermore, as an adaptor protein, SMAD7 antagonizes inhibitory intramolecular interactions between the SMURF2 C2 and HECT domains and relieves C2 domain-dependent autoinhibition, ultimately promoting SMURF2 activity. Besides ubiquitin-mediated regulation of receptors, SMAD7-SMURF2 complexes were recently shown to promote proteasomal degradation of SMAD anchor for receptor activation (SARA), a positive regulator of TGF-β signaling (Wojtowicz et al. Citation2020). Lastly, the SMAD7 PY motif is involved in binding Yes-Associated Protein (YAP)65 in order to co-repress TGF-β signaling (Ferrigno et al. Citation2002).
Contrary to the MH2 domain, the N-terminal region of SMAD7 (amino acids 1-260) shows limited similarity with the MH1 domain found in SMAD1 to SMAD5. The SMAD7 N-terminal region has several distinct functions in TGF-β signaling. First, the N-terminal amino acid residues 32-90 were found to be important determinants of inhibitory effect on the receptor signaling. Additionally, even though the SMAD7–TβRI interaction is mainly MH2-mediated, the N-terminal region co-determines the affinity of SMAD7 for TβRI. Furthermore, inhibition of TGF-β signaling is potentiated by physical interaction between the SMAD7 N-terminal region and MH2 domain (Hanyu et al. Citation2001). In addition, the N-terminal domain was shown to determine subcellular localization of SMAD7 in multiple cell lines (Itoh et al. Citation1998; Hanyu et al. Citation2001). Furthermore, the N-terminal region of SMAD7 contains a Leu-rich motif (LRM), which is important in the recruitment of the E2 ubiquitin-conjugating enzyme UbcH7 to the SMURF2 HECT domain (Ogunjimi et al. Citation2005). Moreover, the SMAD7 N-terminal region has been suggested to contain DNA-binding ability. In R-SMADs, the MH1 domain is responsible for DNA-binding activity. The N-terminal region of I-SMADs, however, lacks the DNA-binding β-hairpin motif. I-SMADs are predicted to contain a zinc atom in their N-terminal region, similar to the zinc atom in R-SMADs that facilitates DNA-binding (Hariharan and Pillai Citation2008), indicating a possible role for the SMAD7 N-terminal region in DNA-binding. Taken together, these studies demonstrate how divergent structural features of SMAD7, as compared to R- and Co-SMADs, facilitate its function.
Regulation of SMAD7 expression and function
SMAD7 is differentially expressed across various tissues and is subject to tight regulation by a range of molecules and a number of regulatory mechanisms. In this section, we will discuss various mechanisms by which SMAD7 expression and function are controlled, including transcriptional, post-transcriptional and post-translational mechanisms ().
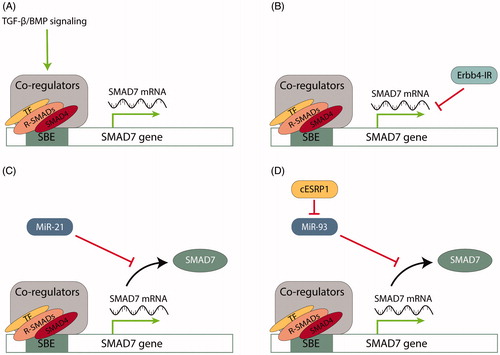
Figure 3. Examples of SMAD7 regulation on a transcriptional and post-transcriptional level. A. TGF-β and BMP signaling activation positively regulate SMAD7 transcription. The SMAD7 promoter is a target for direct association with R- and Co-SMADs, stimulating promoter activity. B. Long non-coding RNA Erbb4-IR suppresses SMAD7 transcription by binding to the 3′ UTR of the SMAD7 gene. C. MiR-21 modulates SMAD7 expression by targeting mRNA for translational repression by pairing to sequence in the mRNA 3′-untranslated region (3′ UTR). D. Circular RNA cESRP1 sponges miR-93, a negative regulator of SMAD7, to inhibit TGF-β signaling. SBE, SMAD binding element; TF, transcription factor.
Transcriptional regulation of SMAD7
SMAD7 mRNA expression is promoted by TGF-β family members (Nakao et al. Citation1997; Afrakhte et al. Citation1998). SMAD3 and SMAD4 directly associate with the SMAD7 promoter, through its consensus SBE, stimulating promoter activity () (Nagarajan et al. Citation1999; Von Gersdorff et al. Citation2000). As SMAD7 functions as a TGF-β signaling antagonist, it thereby participates in a negative feedback circuit.
Gene expression can also be modulated by controlling the accessibility of transcription factors to the corresponding gene through covalent histone modification (Strahl and Allis Citation2000). Histone methylation and acetylation are reversible and their effects are context dependent. Histone deacetylases (HDACs) regulate the acetylation of histone proteins, for the purpose of gene expression control. TGF-β stimulation has been shown to increase histone H4 acetylation at the SMAD7 promoter 3.2-fold. This increase corresponded with an induction of SMAD7 mRNA (Alliston et al. Citation2005). Corepressor Evi-1 and associated corepressor C-terminal-binding protein (CtBP) obstructed TGF-β-induced histone acetylation at the endogenous SMAD7 promoter, mitigating SMAD7 mRNA expression. In a similar fashion, transcription co-factor Ski, in cooperation with protein arginine methyltransferase 5 (PRMT5) and HDAC3, is present at the SMAD7 promoter and promotes SMAD7 transcriptional repression (Tabata et al. Citation2009), suggesting a role for PRMT5 and HDAC3 in maintaining the basal repressed state of SMAD7.
Noncoding RNA-mediated regulation of SMAD7 expression
SMAD7 expression is extensively regulated at the RNA level by noncoding RNAs (ncRNAs), such as microRNAs (miRNAs), circular RNAs (circRNAs) and long noncoding RNAs (lncRNAs). MiRNAs are small, endogenous non-coding RNA molecules of approximately 22 nucleotides in length, involved in the regulation of gene expression (Bartel Citation2004; Ambros Citation2004). In the nucleus, RNA polymerase II or III transcribes primary miRNA (pri-miRNA). Subsequently, 70 nucleotide stem-loop precursor miRNAs (pre-miRNAs) are excised by the RNase III DROSHA microprocessor complex and exported to the cytoplasm (Kim et al. Citation2009). The cytoplasmic RNase III enzyme Dicer completes the processing of pre-miRNAs to form mature miRNAs. MiRNAs modulate expression by targeting mRNA for translational repression by pairing to sequences in the mRNA 3′-untranslated region (3′ UTR) in target mRNAs or inhibiting translation (Fabian et al. Citation2010). Hundreds of mammalian miRNAs have been identified as key post-transcriptional regulators in biological processes such as development, cell proliferation and cell death (Landgraf et al. Citation2007).
MiRNA processing is promoted by TGF-β and BMP signaling (Davis et al. Citation2010). Following ligand stimulation, R-SMADs associate with the RNA SMAD-binding element (R-SBE) of pri-miRNA transcripts in order to facilitate DROSHA-mediated miRNA maturation. This indicates that R-SMADs modulate gene expression by (A) transcriptional induction and (B) facilitating post-transcriptional processing of miRNAs. Here, we provide an overview of several key miRNAs that regulate SMAD7 expression, and discuss other examples of ncRNA-mediated SMAD7 regulation.
MiR-21
One of the most extensively studied miRNA regulating SMAD7 is human miR-21 (hsa-miR-21). Its role in regulating SMAD7 expression has mainly been examined in the context of disease. MiR-21 was first identified as a regulator of SMAD7 expression in liver fibrosis based on TargetScan predictions (Marquez et al. Citation2010). Marquez et al. established post-transcriptional regulation of SMAD7 by miR-21. They demonstrated direct interaction between miR-21 and the SMAD7 3’UTR using site-directed mutagenesis in the miR-21 seed sequence, which is the primary determinant of miRNA specificity (Marquez et al. Citation2010). Afterwards, it was confirmed that treatment with miR-21 precursor indeed leads to decreased SMAD7 protein levels in fibroblasts (Liu et al. Citation2010).
MiR-21 has been extensively studied in a range of diseases, such as cancer, cardiovascular disease, and pulmonary disease. In malignancies, miR-21 is a central player as an oncogenic miRNA (oncomiR) targeting tumor suppressor proteins, including for example PTEN (Feng and Tsao Citation2016). MiR-21 is upregulated in intraductal epithelial proliferations of breast tissue compared to healthy tissue (Chen et al. Citation2013). This overexpression correlates with decreased SMAD7 levels. MiR-21 knockdown restored SMAD7 mRNA levels in MCF-7 and Hs578T breast cancer cells. Likewise, SMAD7 mRNA expression was significantly increased in miR-21 knockout HeLa cells (Chen et al. Citation2015). Wang et al. demonstrated that SMAD7 is also a direct target for transactivated miR-21 in colorectal cancer (Wang et al. Citation2017). Similar results have been reported for coronary heart disease, autoimmune disease, diabetes and during wound healing (Li et al. Citation2015, Citation2018; Murugaiyan et al. Citation2015; Luo et al. Citation2017).
Chang et al. have demonstrated a divergent role for hsa-miR-21-5p in the regulation of SMAD7 in hypoxic conditions, compared to normoxic conditions. Here, 5p refers to the 5p strand that is located in the forward (5′-3′) position, whereas the 3p strand is present in the reverse position. In hypoxia, hsa-miR-21-5p expression increased protein expression of SMAD7 in human umbilical vein endothelial cells, whereas at normoxia hsa-miR-21-5p induces downregulation of SMAD7 in these cells, providing a prime example of the effect of context-dependency on functional output (Chang et al. Citation2017). In conclusion, the fact that SMAD7 is a confirmed target for miR-21 in a plethora of diseases highlights the importance of SMAD7 in maintaining appropriate TGF-β signaling.
SMAD7 regulation by diverse miRNAs
MiRNA-mediated regulation of SMAD7 is not limited to miR-21. The miR-15 family, consisting of six highly conserved miRNAs (miR-15a/b, miR-16, miR-195, miR-497, miR-322), inhibits TGF-β signaling in cardiovascular tissues (Tijsen et al. Citation2014). Knockdown of the miR-15 family resulted in upregulated SMAD7 mRNA levels and increased SMAD7 3′UTR reporter activity in cardiomyocytes. It remains unclear, however, whether these effects are due to direct interaction between miR-15 family members and SMAD7 3’UTR (Tijsen et al. Citation2014). Direct regulation of SMAD7 by miR-15b was demonstrated in osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) (Fang et al. Citation2019).
Interestingly, distinct miRNA variants derived from the same precursor can affect SMAD7 gene expression in different cell types. For example, from the miR-181 precursor four mature miRNAs can be produced: miR-181a, miR-181b, miR-181c or miR-181d, of which MiR-181a was found to induce repression of its functional target SMAD7 in ovarian cancer cells to promote TGF-β-mediated epithelial-to-mesenchymal transition (EMT) (Parikh et al. Citation2014), whereas MiR-181c downregulated SMAD7 expression in neuroblastoma and osteosarcoma (Li et al. Citation2014; Fu et al. Citation2019). Additionally, miR-181 variants have been shown to regulate T cell phenotype in autoimmune neuroinflammation by directly targeting SMAD7 mRNA (Ghorbani et al. Citation2017; Zhang et al. Citation2018).
Many other miRNAs have been shown to regulate SMAD7 expression (). Of note, in many cases of miRNA-mediated SMAD7 regulation it remains unclear which variant governs the regulation, and whether the repression of SMAD7 expression occurs due to direct interaction with the SMAD7 3’UTR or through intermediary regulators, such as SMAD7 interacting proteins.
Table 1. MiRNAs regulating SMAD7 expression in the context of diseasea.
SMAD7 regulation by other types of ncRNAs
In addition to miRNA-mediated regulation, SMAD7 is subject to regulation by circRNAs, and lncRNAs. CircRNAs, which result from backsplicing of pre-mRNAs, regulate the expression of protein-coding transcripts (Huang et al. Citation2017). CircRNAs can function as a sponge for miRNAs, obstructing their regulatory function. In TGF-β signaling, circRNA cESRP1 protects SMAD7 from downregulation by sponging miR-93-5p (Huang et al. Citation2020). Furthermore, Circ-LARP4 has been found to positively regulate SMAD7 expression in non-small-cell lung cancer (Shi et al. Citation2020).
Finally, lncRNAs have also been attributed with SMAD7 regulatory functions. For example, linc-SMAD7 increases SMAD7 expression by sponging SMAD7-targeting miR-125b (Song et al. Citation2018). Other instances in which lncRNAs were found to influence SMAD7 expression are summarized in .
Table 2. LncRNA that positively and negatively regulate SMAD7.
Control of SMAD7 by post-translational modifications
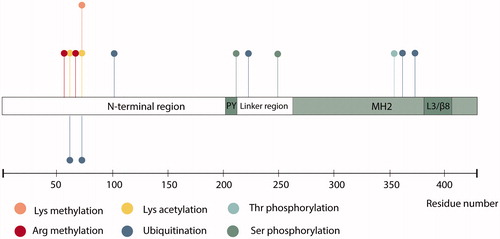
SMAD7 is also known to be extensively regulated by post-translational modifications (PTMs) such as methylation, ubiquitination, acetylation and phosphorylation (), which are discussed in more detail below (Xu et al. Citation2016).
Figure 4. Schematic representation of SMAD7 including identified post-translational modifications. PY, proline-tyrosine motif; MH2, Mad homology 2 domain; L3/β8, L3/β8 loop-strand module.
Methylation
Methyltransferases are able to transfer methyl groups to histones, but are also able to target a variety of non-histone proteins, including SMAD7. Histone-lysine N-methyltransferase SET9 promotes TGF-β signaling by methylating SMAD7. This methylation of SMAD7 significantly increased the interaction with E3 ubiquitin ligase Arkadia (also termed RNF111), facilitating SMAD7 ubiquitination and proteasomal degradation (Elkouris et al. Citation2016). SMAD7 is also a substrate for PRMT1, another methyltransferase (Inamitsu et al. Citation2006; Katsuno et al. Citation2018). Arg-57 and Arg-67 methylation of SMAD7 by PRMT1 led to lowered TβRI-binding efficiency and increased degradation of SMAD7. Taken together, these results indicate that methylation of the SMAD7 protein promotes its degradation.
Phosphorylation
In R-SMADs and SMAD4, the linker region is a target for phosphorylation by a large variety of serine/threonine kinases such as mitogen-activated protein kinases (MAPKs), extracellular-signal regulated kinase (ERK), Jun N-terminal kinase (JNK) and p38 kinase (p38) (Kamato et al. Citation2013). Linker region phosphorylation of R-SMADs regulates SMAD stability, activity and nuclear transport (Matsuzaki Citation2011). SMAD7 is subject to serine phosphorylation in the linker region, at S249 (). Phosphorylation at this site does not inhibit SMAD7 regulatory function in TGF-β or BMP signaling, but does affect SMAD7-dependent transcriptional activity (Pulaski et al. Citation2001). Conversely, linker region phosphorylation at S206, directly adjacent to the PY motif, increases binding affinity between SMAD7 and the WW4 domain of E3 ubiquitin ligase WWP2 (Wahl et al. Citation2019). Besides serine phosphorylation, SMAD7 can also become phosphorylated at T354 (Casado et al. Citation2013).
Ubiquitination
Ubiquitination (also called ubiquitylation) is a crucial regulator of protein stability. This reversible protein modification occurs through an enzymatic cascade performed in succession by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s). The key regulatory roles of E3 ligases in TGF-β signaling have been previously reviewed (Imamura et al. Citation2013). Here, we highlight two vital E3s that regulate SMAD7 stability. Arkadia is an E3 ligase that promotes TGF-β signaling by inducing polyubiquitination and degradation of SMAD7 (Koinuma et al. Citation2003). E3 ubiquitin ligase atrophin 1-interacting protein 4 (AIP4) (also termed ITCH) was shown to positively regulate TGF-β signaling via SMAD7 ubiquitination and subsequent degradation (Park et al. Citation2015). However, AIP4 is also able to affect SMAD7 function independent of its catalytic activity. A catalytic mutant of AIP4 stabilizes the SMAD7-TβRI complex, effectuating TGF-β signaling inhibition (Lallemand et al. Citation2005). Furthermore, in an extensive mass spectrometry experiment, SMAD7 was found to be ubiquitinated at K101. However, the functional effect of ubiquitination at this site remains unknown (Akimov et al. Citation2018).
An important SMAD7 function is its role as adaptor for E3 ligase SMURF2. SMAD7 recruits SMURF2 to the cell membrane in order to induce TβRI degradation by means of ubiquitin-mediated proteolysis (Kavsak et al. Citation2000). Conflicting results have been described on whether SMAD7 itself is a target for ubiquitination by SMURF2. Kavsak et al. showed increased SMAD7 degradation upon overexpression of SMURF2 (Kavsak et al. Citation2000). In contrast, Zhang et al. report physical interaction between SMURF2 and SMAD7 but no evidence for SMURF2-mediated ubiquitination and degradation of SMAD7 (Zhang et al. Citation2001). More recent reports, however, again suggest a role for SMURF2 in SMAD7 proteasomal degradation (Zhao et al. Citation2016; Liu et al. Citation2017).
Deubiquitination
Emerging players in the field of SMAD7 regulation are deubiquitinating enzymes (DUBs). In 2004, associated molecule with the SH3 domain of STAM (AMSH2) was discovered to interact with SMAD7 and to positively regulate TGF-β signaling (Ibarrola et al. Citation2004). Moreover, CYLD deubiquitinates SMAD7 at K360 and K374 and thereby negatively regulates TGF-β-induced TAK1 kinase activity in CD4+ T cells (Zhao et al. Citation2011). Ubiquitin-Specific Peptidase 26 (USP26) has been found to deubiquitinate and thereby stabilize SMAD7, resulting in reduced TGF-β signaling (Kit Leng Lui et al. Citation2017). Similarly, OTU Deubiquitinase 1 (OTUD1) cleaves polyubiquitin chains from SMAD7 K220, exposing the PY motif. This in turn enables SMURF2 binding and promotes receptor turnover (Zhang et al. Citation2017).
Acetylation
SMAD7 stability is regulated by acetylation and the consequential competition between this modification and ubiquitination (Grönroos et al. Citation2002). SMAD7 interaction with transcription coactivator and acetyltransferase p300 leads to its acetylation at two N-terminal lysines (K64 and K70), preventing ubiquitination at these sites. Additionally, HDAC1-mediated deacetylation of SMAD7 decreases SMAD7 stability by enhancing its ubiquitination and subsequent degradation in the proteasome (Simonsson et al. Citation2005). In accordance with these findings, Monteleone et al. demonstrated that SMAD7 is not transcriptionally regulated in the human gut. Rather, SMAD7 is post-translationally stabilized by p300-mediated acetylation (Monteleone et al. Citation2005). Taken together, these results suggest that SMAD7 stability is regulated by acetylation and ubiquitination in a mutually exclusive manner.
Protein interactions govern SMAD7 stability and activity
Another manner of SMAD7 regulation is via protein-protein interactions. E3 ubiquitin ligase SMURF1 directly binds SMAD7 and facilitates cytoplasmic localization of SMAD7, targeting SMAD7 to the plasma membrane through its N-terminal C2 domain. This enables negative regulation of TGF-β signaling independent of its catalytic function (Suzuki et al. Citation2002). SMAD7 can also be regulated by direct interaction with other proteins, such as Vpr binding protein (VprBP) and REGγ. VprBP was found to interact with SMAD7 to promote SMAD7-SMURF1-TβRI complex formation (Y et al. Citation2020). Conversely, REGγ, a member of the 11S proteasome activators, binds to SMAD7 in order to promote its turnover (Jiao et al. Citation2020a). The microtubule actin crosslinking factor 1 (MACF1), a protein highly abundant in bone tissue, has been identified as a SMAD7 interaction partner in mesenchymal stem cells. MACF1 facilitates SMAD7 nuclear translocation to promote osteogenic differentiation (Zhao et al. Citation2020).
As discussed, SMAD7 requires interaction with other proteins in order to successfully antagonize TGF-β and BMP signaling. For instance, SMAD7 recruits E3 ubiquitin ligases, such as NEDD4L and SMURF1/2 to induce receptor polyubiquitination and degradative endocytosis (Kavsak et al. Citation2000; Ebisawa et al. Citation2001; Kuratomi et al. Citation2005). SMAD7 activity can, therefore, be regulated by competition for its binding partners. For example, SMURF2 establishes mutually exclusive complexes with SMAD7 or RNF11 (Malonis et al. Citation2017). Accordingly, RNF11 regulates SMAD7 activity by sequestering SMURF2 at the cell membrane. Moreover, c-SRC-mediated phosphorylation of SMURF2 at W14 and W434 was found to reduce SMAD7 binding, enhance intramolecular interactions within SMURF2, and result in inhibition of SMURF2 activity (Sim et al. Citation2019).
In addition to being a target for deubiquitination, several DUBs interact with SMAD7 to regulate the TGF-β signaling pathway (Herhaus and Sapkota Citation2014). Both USP11 and USP15 are able to bind to SMAD7 to deubiquitinate and stabilize the TβRI, enhancing signaling TGF-β signaling (Al-Salihi et al. Citation2012; Eichhorn et al. Citation2012). Moreover, SMAD7 recruits ubiquitin C-terminal hydrolase 37 (UCH37) to antagonize the SMURF-mediated ubiquitination and subsequent degradation of the TβRI (Wicks et al. Citation2005; Cutts et al. Citation2011). Taken together, these studies indicate that DUBs can counteract SMAD7 function at the receptor level.
In conclusion, SMAD7 functioning is in part dependent on interaction with interaction partners, such as SMURF1/2 and is affected by competition for and modification of these interaction partners.
SMAD7 as signaling integrator
At any given moment, cells receive a multitude of signals simultaneously that are integrated to determine cell shape and function. A single pathway is often connected at different steps with other pathways, resulting in complicated signaling networks that allow for appropriate spatio-temporal responses. SMAD7 is an important regulatory TGF-β signaling component by which signaling crosstalk is achieved. Initially identified in endothelial cells as a shear stress-inducible gene (Topper et al. Citation1997), SMAD7 is a potent antagonist of TGF-β and BMP signaling. SMAD7 expression is induced by TGF-β and BMP stimulation, and thereby participates in a negative feedback loop controlling the duration and intensity of the TGF-β family signaling response (Nakao et al. Citation1997). Efficient induction of SMAD7 by TGF-β, however, depends on various transcription factors and cofactors, such as activator protein 1 (AP-1), stimulating protein-1 (Sp1), transcription factor mE3 (TFE-3) , activating transcription factor 2 (ATF2), CBP/p300 and Forkhead box H1 (FOXH1) (Brodin et al. Citation2000; Hua et al. Citation2000; Uchida et al. Citation2001; Jungert et al. Citation2006; Gohla et al. Citation2008). Many of these factors are regulated by other signaling cascades besides TGF-β as well. Moreover, SMAD7 expression can be induced by epidermal growth factor (EGF), human epidermal growth factor receptor 2 (HER2/Neu), UV irradiation, and TPA (Afrakhte et al. Citation1998; Quan et al. Citation2001; Dowdy et al. Citation2003; Tsunobuchi et al. Citation2004). Thus, this suggests a role for SMAD7 as integrator and crosstalk mediator between TGF-β signaling and other signaling pathways. Here, we provide a comprehensive overview of SMAD7 as a signaling integrator.
EGFR signaling and the MAPK/ERK pathways
Crosstalk between TGF-β and other signaling pathways was originally demonstrated by the discovery that EGF increased the expression of SMAD7 mRNA (Afrakhte et al. Citation1998). The EGF receptor (EGFR) belongs to the receptor tyrosine kinase (RTK) family and is activated by various ligands, including EGF and TGF-α. Luwor et al. reported that EGFR-induced desensitization to TGF-β is dependent on signal transducer and activator of transcription (STAT)3-mediated SMAD7 promoter activation (Luwor et al. Citation2013). On the other hand, SMAD7 regulates EGF signaling as well. SMAD7 sequesters E3 ubiquitin ligase c-Cbl, thereby inhibiting ligand-induced EGFR ubiquitination and proteasomal degradation in human HaCaT keratinocytes (Ha Thi et al. Citation2015). Similarly, SMAD7 overexpression stimulates EGFR-signaling activation and its downstream signaling in skin carcinogenesis (Ha Thi et al. Citation2019).
Mitogen-activated protein kinases (MAPKs), including ERK1/2, JNK1/2/3, and p38/MAPKs, are serine–threonine kinases and essential mediators of intracellular signaling events involved in regulating cellular processes related to proliferation, differentiation, survival, and death in response to a diverse array of stimuli. Extracellular stimuli, such as EGF, can prompt a phosphorylation cascade from MAP kinase kinase kinase (MAPKKK) to MAP kinase kinase (MAPKK), and ultimately MAPK (Pearson et al. Citation2001). ERK1/2, JNK1/2/3, and p38 MAPK have all been implicated in the transcriptional regulation of SMAD7 () (Brodin et al. Citation2000; Uchida et al. Citation2001; Dowdy et al. Citation2003). In turn, SMAD7 itself regulates MAPK signaling. Transient and stable SMAD7 expression leads to sustained JNK activation, potentiating apoptosis (Mazars et al. Citation2001). Moreover, SMAD7 expression is critical for TGF-β-induced activation of MKK3 and p38 kinases (Edlund et al. Citation2003). Thus, SMAD7 expression is regulated by EGFR and all MAPK branches and, in turn, SMAD7 influences EGFR and MAPK signaling.
Figure 5. SMAD7 is regulated by and regulates various signaling pathways. In the Wnt signaling pathway, SMAD7 regulates the activity and stability of β-catenin by functioning as a SMURF2 adaptor. Similarly, SMAD7 can affect NFκB signaling by inhibiting phosphorylation and degradation of IκB and disrupting recruitment of the TAK1 complex to TRAF2. SMAD7 expression can be positively regulated by inflammatory cytokines such as TNF-α and IFN-γ, and YAP/TAZ, effectors in the Hippo signaling pathway. Lastly, SMAD7 expression is positively regulated by laminar shear stress, UV light, TPA and EGF. SMAD7 is also important for JNK and p38 activation. ERK, Extracellular-signal-regulated kinase; JAK, Janus kinase; JNK, c-Jun N-terminal kinase; NFκB, Nuclear factor kappa B; STAT, Signal transducer and activator of transcription; TAK, TGF-β-activated kinase; TAZ, Tafazzin; TRAF, TNF receptor-associated factor; YAP, Yes-associated protein.
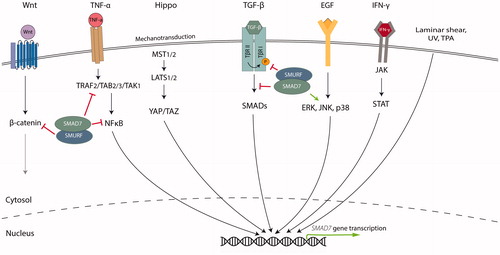
The IL, TNF, and IFN-γ pathways
Interleukines (ILs), tumor necrosis factor-alpha (TNF-α), and interferon-gamma (IFN-γ) are secreted factors that regulate important immune responses, by signaling through the NF-κB and just another kinase (JAK)/STAT pathways in receiving cells. Upon IL- or IFN-γ receptor binding, JAK becomes activated by transphosphorylation and in turn phosphorylates STAT proteins on tyrosine residues. Subsequently, STAT proteins disassociate from the receptor, dimerize and translocate into the nucleus (Rawlings et al. Citation2004). TNFα can trigger activation of NF-κB proteins, including p50, p52, RelA/p65, and RelB, by inducing degradation of the IκB protein. After entering the nucleus, these activated NF-κB proteins function as transcription factors () (Napetschnig and Wu Citation2013).
Crosstalk between SMAD7 and the IFN-γ/STAT pathway
The TGF-β and IFN-γ signaling pathways often have distinct opposite effects and reciprocally inhibit each other, for example in the regulation of inflammatory responses. Through JAK1 and STAT1, IFN-γ induces the expression of SMAD7, thereby preventing the interaction of SMAD3 with the TβRI (Ulloa et al. Citation1999). Further investigation of IFN-γ-mediated reduction of TGF-β signaling showed that Y-box protein-1 (YB-1) activation by JAK1 is necessary for IFN-γ-mediated upregulation of SMAD7 (Dooley et al. Citation2006). YB-1 binds directly to a recognition site in the SMAD7 promoter to induce transcription. Moreover, IL-6 treatment results in STAT3-dependent transcriptional induction of SMAD7 in gp130Y757F/Y757F mice (Jenkins et al. Citation2005).
Crosstalk between SMAD7 and TNF-α/NF-κB signaling
The proinflammatory cytokines TNF-α and IL-1β induce SMAD7 expression in an NF-κB/RelA-dependent manner in mouse fibroblasts (Bitzer et al. Citation2000). However, activation of NF-κB by TNF-α stimulation was shown to inhibit the SMAD7 promoter in HEK293 cells. This inhibition could be reversed by overexpression of the transcription co-activator p300, indicating that NF-κB regulates SMAD7 expression by competition for the common transcription coactivators (Nagarajan et al. Citation2000). How SMAD7 transcription is modulated by NF-κB might therefore be dependent on the cellular context. Transmembrane protein CD40, member of the TNF-receptor superfamily, promotes SMAD7 expression through the NF-κB pathway (Patil et al. Citation2000). Conversely, SMAD7 has been reported to have an anti-inflammatory effect by affecting NF-κB signaling. SMAD7 could promote expression of IκB, a specific inhibitor of NF-κB (Wang et al. Citation2005). Similarly, in colon cancer cells, SMAD7 can inhibit the phosphorylation and degradation of IκB (Grau et al. Citation2006). Another mechanism through which SMAD7 can regulate NF-κB signaling is by formation of SMAD7-TGF-β-activated kinase (TAK1)-binding protein (TAB)2 and SMAD7-TAB3 complexes, disrupting recruitment of the TAK1 complex to TNF receptor–associated factor 2 (TRAF2), and thereby blocking propagation of proinflammatory signals (; Hong et al. Citation2007).
Wnt signaling
The Wnt signaling pathway initiates when a Wnt-protein ligand binds to a Frizzled family receptor, triggering intracellular signaling cascades via the Disheveled protein. Canonical Wnt signaling leads to transcriptional regulation of target genes via the nuclear translocation of β-catenin, an essential mediator of this pathway (Clevers and Nusse Citation2012). Edlund et al. demonstrated physical interaction between SMAD7 and β-catenin (Edlund et al. Citation2005). Additionally, SMAD7 has been shown to bind β-catenin and thereby promote its degradation through SMURF2-mediated ubiquitination (Han et al. Citation2006). Axin, a key component of the canonical Wnt pathway, suppresses Wnt signal transduction by promoting β-catenin degradation. SMAD7 can bind to Axin in order to stabilize β-catenin (Tang et al. Citation2008). Conversely, Axin can function as a scaffold protein in TGF-β signaling by formation of a multimeric complex with SMAD7 and Arkadia. This promotes SMAD7 turnover via proteasomal degradation (Liu et al. Citation2006). Lastly, Yanagida et al. found that supplementing exogenous Dickkopf-related protein (Dkk), a Wnt antagonist, increased SMAD7 expression in a liver fibrosis mice model (Yanagida et al. Citation2011). These results indicate an intimate relationship between SMAD7 and Wnt/β-catenin signaling.
Hippo signaling
The evolutionarily conserved Hippo pathway is characterized by a kinase cascade, where, in mammals, Macrophage-stimulating protein (MST)1 and 2 and scaffold protein SAV1 form a complex in order to phosphorylate and activate Large tumor suppressor kinase 1 (LATS)1 and 2. In turn, LATS1 and 2 phosphorylate and inhibit two important downstream effectors of the Hippo pathway; Yes-associated protein (YAP) and Tafazzin (TAZ) (Meng et al. Citation2016). YAP is able to enhance TGF-β signaling inhibition by facilitating recruitment of SMAD7 to the TβRI (Ferrigno et al. Citation2002). In human skin dermal fibroblasts, YAP/TAZ have been found to inhibit TGF-β signaling by promoting SMAD7 expression via transcription factor AP-1 (Qin et al. Citation2018). Moreover, loss of tumor suppressor Merlin, an upstream activator of the Hippo signaling pathway, leads to unrestrained YAP/TAZ activity and is concordant with decrease of SMAD7 expression (Mota et al. Citation2018).
Notch signaling
The Notch signaling pathway is important for regulation of cell differentiation, proliferation, and cell death. The Notch receptor is a cell-surface receptor that transduces signals by interacting with transmembrane ligands, such as Delta-like and Jagged, on juxtaposed cells (Siebel and Lendahl Citation2017). Tsai et al. determined that SMAD7 is a Notch signaling target in limbal epithelial stem cells, and propose that it may facilitate the function of Notch in mitigating EMT (Tsai et al. Citation2014). Additionally, Notch signaling activation was found to be essential for maintaining vascular endothelial growth factor (VEGF)165-induced SMAD7 expression in endometrial stromal cells (Lv et al. Citation2019).
The ATM-dependent DNA damage signaling pathway
As with ultraviolet (UV) light, radiation therapy can induce SMAD7 mRNA (Kruse et al. Citation2009). Upon TGF- β stimulation, SMAD7 is able to affect the ataxia-telangiectasia mutated (ATM)-dependent DNA damage pathway by acting as a scaffold protein to facilitate interaction between p38 MAPK, ATM and p53, thereby promoting their activation. SMAD7 was found to colocalize with H2A histone family member X (γH2AX) in DNA damage foci. Moreover, SMAD7 is necessary for proper TGF-β induced cell cycle arrest (Zhang et al. Citation2006). Consistent with these results, Park et al. demonstrated that SMAD7 promotes the interaction between ATM and Nibrin (NBN, also termed NBS1), enhancing DNA repair (Park et al. Citation2015). Furthermore, SMAD7 localizes to irradiation-induced double strand breaks (Wang et al. Citation2013). These results indicate a role for SMAD7 in facilitating DNA damage repair.
In summary, SMAD7 is subject to regulation by a myriad of pathways, integrating information from many extracellular stimuli (). Accordingly, targeting SMAD7 for therapeutic intervention might produce undesirable outcomes in patients, because of the high degree of connectivity between pathways and the integral role of SMAD7 in a multitude of ways.
Physiological roles of SMAD7 in health and disease
Since the discovery of SMAD7, much research has been conducted to uncover its biological functions in the context of human diseases. In this section, we discuss what genetic studies, in mice and humans (if available), and recent biochemical studies have revealed about the physiological role of SMAD7 in fibrosis, cancer, and inflammatory bowel disease. We also discuss opportunities for therapeutic intervention by targeting SMAD7.
Fibrosis
Genetic studies in mice
TGF-β signaling is an important signaling pathway in tissue fibrosis (Hu et al. Citation2018). Across numerous fibrosis models, expression or disruption of SMAD7 resulted in reduced or enhanced fibrosis in mice respectively (Nakao et al. Citation1999; Dooley et al. Citation2008; Hamzavi et al. Citation2008; Chung et al. Citation2009; He et al. Citation2009). Pancreatic fibrosis is a characteristic hallmark of chronic pancreatitis. Upon induction of chronic pancreatitis, mice with a deficiency in exon I of SMAD7 (SMAD7ΔE1) showed a more severe response characterized by an enhanced accumulation of extracellular matrix (ECM), an increased number of mesenchymal cells, and enhanced infiltration of inflammatory cells (Li et al. Citation2016). Taken together, these mice studies suggest a protective function of SMAD7 in fibrosis in various organs through inhibition of profibrogenic TGF-β signaling.
Human biological studies
Recent research using human endometrial tissue supports the view of SMAD7 as a protective factor in fibrogenesis. It was reported that SMAD7 and Notch are essential downstream molecules for VEGF165 functioning in endometrial fibrosis. VEGF165 reverses TGFβ1-stimulated fibrotic gene expression. However, this protective effect is SMAD7-dependent (Lv et al. Citation2019). Collectively, these recent studies provide evidence for the perceived protective role for SMAD7 in fibrotic disease.
Cancer
Genetic studies in mice
The role of SMAD7 in cancer growth and progression has been previously reviewed (Stolfi et al. Citation2013). TGF-β signaling has a dual role in tumor initiation and progression. In early stages, TGF-β signaling functions as a tumor suppressor. However, in later stages aberrant TGF-β signaling can facilitate EMT, migration, invasion and metastasis (Massagué Citation2008). In a mouse model for squamous cell carcinoma (SCC), SMAD7 enhanced keratin 8, a marker of malignant conversion (Liu et al. Citation2003). Contrarily, transgenic expression of SMAD7 was found to reduce 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced hyperplasia in mouse skin (Hong et al. Citation2007). 1205Lu melanoma cells with stable SMAD7 overexpression showed diminished ability to form tumors by xenograft experiment in nude mice (Javelaud et al. Citation2005). Moreover, these mice demonstrated prolonged survival and significantly less bone metastasis (Javelaud et al. Citation2007). Additionally, SMAD7wt transgenic mice showed significantly greater papilloma formation, compared to control and SMAD7mut mice (Ha Thi et al. Citation2019), while conditional SMAD7 deletion led to highly proliferative and invasive cells and sustained melanoma growth and metastasis (Tuncer et al. Citation2019). Taken together, these contrary results highlight the complicated, context-dependent role of both SMAD7 and the TGF-β pathway in cancer.
SMAD7 overexpression has been shown to inhibit carcinogenesis and metastasis under certain conditions. In murine osteosarcoma and breast cancer models, SMAD7 overexpression resulted in a decrease in metastasis and longer survival, compared to control mice (Azuma et al. Citation2005; Lamora et al. Citation2014). Wang et al. showed that SMAD7 KO mice were more susceptible to N-nitrosodiethylamine (DEN)-induced hepatocellular carcinoma (HCC) than their wild-type counterparts (Wang et al. Citation2013). Adenoviral overexpression of SMAD7 in SCC cells inhibited growth of xenografts in severe combined immunodeficient (SCID) mice (Leivonen et al. Citation2006). Similarly, SMAD7 expression in CD4+ T cells was found to prevent colitis-associated colorectal cancer due to increased expression of IFNγ (Rizzo et al. Citation2011). Even independent of chronic inflammation, T-cell-specific overexpression of SMAD7-protected mice against tumor development (Rizzo et al. Citation2014).
Conversely, SMAD7 is also able to promote cancer initiation and progression. It has been reported that SMAD7 overexpression in colon adenocarcinoma cells induces tumorigenicity (Halder et al. Citation2005). Knockdown of SMAD7 using antisense oligonucleotide resulted in reduced tumor growth in RAG1-/- mice (Stolfi et al. Citation2014). Moreover, SMAD7 promotes hepatic metastasis in CRC (Halder et al. Citation2008). Transgenic pancreas-specific expression of SMAD7 induced premalignant ductal lesions in mice (Kuang et al. Citation2006). Hepatocyte specific deletion of SMAD7 revealed that SMAD7 inhibits IL-6 production in hepatocellular carcinoma, thereby decreasing STAT3 signaling and accelerating tumor growth (Feng et al. Citation2017). In bronchiolar exocrine cells, present in the mouse airway, constitutively expressed SMAD7 increased incidence of urethane-induced lung cancer (Luo et al. Citation2010). Altogether, these studies illustrate a complex, multifaceted role of SMAD7 in tumorigenesis.
Genetic studies in humans
In 2007, researchers first demonstrated the importance of SMAD7 in colorectal cancer (CRC) in a genome-wide association study (GWAS) (Broderick et al. Citation2007). They showed significant association between single nucleotide polymorphism (SNP) rs4939827 of SMAD7 and CRC. Two other SNPs (rs4464148 and rs12953717) were shown to confer modest susceptibility to CRC (Tenesa et al. Citation2008). In 2014, Fortini et al. discovered four SMAD7 NSPs (rs6507874, rs6507875, rs8085824, and rs58920878) that were in linkage disequilibrium with rs4939827 (Fortini et al. Citation2014). In normal colon tissues, the SNP rs8085824 C (minor) allele was found to correlate with increased SMAD7 expression. The discovery of these SNPs sparked a new line of investigation into the pathogenesis of CRC, however the functional significance of SMAD7 variants is not yet clear and provides avenues for future research.
Human biological studies
As in in vivo experiments, in vitro research demonstrates a dual role for SMAD7 in cancer initiation and progression. SMAD7 is commonly upregulated in CRC tissue compared to healthy adjacent tissue and SMAD7 inhibition causes a block in the S phase of the cell cycle, resulting in reduced CRC cell growth in vitro (Stolfi et al. Citation2014). It was discovered that SMAD7 inhibition promotes phosphorylation and activation of the eukaryotic Initiation Factor 2α (eIF2α) pathway via serine–threonine protein kinase RNA (PKR), leading to enhanced cell death (De Simone et al. Citation2017).
Conversely, Krüppel-like factor 4 (KLF4) overexpression inhibits TGF-β-induced cell migration and invasion by enhancing SMAD7 transcription in HCC cells (Sun et al. Citation2017). In glioblastoma (GBM), HECT-domain ubiquitin ligase HERC3 induces ubiquitination-mediated degradation of SMAD7, resulting in TGF-β pathway activation. HERC3-induced downregulation of SMAD7 potentiated TGF-β-induced EMT and was linked to temozolomide chemoradiotherapy resistance in GBM cells (Li et al. Citation2019). Furthermore, it has been reported that REGγ, the gamma subunit of the 11S regulator, regulates EMT, by mediating ubiquitin-independent degradation of SMAD7 in human lung cancer cells (Tong et al. Citation2020). Moreover, REGγ has been found to mediate anaplastic thyroid cancer (ATC)-associated dedifferentiation via SMAD7 degradation (Jiao et al. Citation2020b).
Taken together, these studies suggest that SMAD7 is critical in several cancer hallmarks such as cell death resistance, EMT and altered cellular identity, as well as drug resistance and provide rationale for use of TGF-β inhibitors in (late stage) cancer treatment. However, careful patient selection is warranted in order to ensure the maximum benefit and limit adverse effects.
Inflammatory bowel disease (IBD)
Genetic studies in mice
Inflammatory bowel disease (IBD) is characterized by aberrant TGF-β signaling (Ihara et al. Citation2017). High expression of SMAD7 in CD4+ T Cells was associated with severe colitis. Recombination activating gene 1 (RAG1)-/- mice reconstituted with SMAD7tg cells displayed more pronounced mucosa hyperplasia and drastic loss of goblet cells (Fantini et al. Citation2009). Moreover, transfer of SMAD7-overexpressing T cells was inversely correlated with aryl hydrocarbon receptor (AhR) expression and IL-22 production (Monteleone et al. Citation2016). Recently, a study reported that a short splice form of CYLD (sCYLD) mediates K63-linked ubiquitination and nuclear translocation of SMAD7 in CD4+ T cells, inhibiting TGF-β signaling. Furthermore, regulatory T (Treg) cells from sCYLD+/SMAD7+ mice failed to prevent the induction of adoptive transfer induced colitis in RAG1-/- mice (Tang et al. Citation2019). In addition, dendritic cell (DC)-specific deletion of SMAD7 enhances TGF-β responsiveness and expression of programmed death ligand (PDL)1/2, and promotes Treg differentiation, mediating intestinal inflammation in mice (Garo et al. Citation2019). Sedda et al. demonstrated a novel reciprocal regulatory mechanism between SMAD7 and class III NAD+-dependent deacetylase SIRT1 in the gut using a SMAD7 transgenic mouse model (Sedda et al. Citation2018), adding further nuance to the regulation of SMAD7 in IBD.
SMAD7 antisense oligonucleotide (AON) treatment in inflammatory bowel disease
For the better part of two decades, the scientific community has followed the development of AON treatment in various diseases, including IBD (Scarozza et al. Citation2019). It was reported that SMAD7 AON treatment could potentially restore the endogenous TGF-β signaling pathway and attenuate IBD (Boirivant et al. Citation2006). In 2014, Celgene Corporation acquired Mongersen (GED-301), a SMAD7 specific AON-containing medication, from Nogra Pharma as a late stage candidate in CD. However, despite encouraging results in pre-clinical and early clinical trials, the phase III clinical trial was prematurely terminated due to lack of efficacy (Ardizzone et al. Citation2016; Sands et al. Citation2019). Interestingly, early clinical trials were designed without a control arm (Monteleone et al. Citation2012; Zorzi et al. Citation2012; Feagan et al. Citation2018). However, as the placebo response rate in induction and maintenance trials in CD trials were reported at 28% (95% CI 24–32%) and 26% (95% CI 24–32%) respectively, this demonstrates that it is crucial to establish appropriate controls to exclude placebo effect (Jairath et al. Citation2017). As CD is a relapsing-remitting disease, it is much more difficult to study than continuous and progressive inflammatory disease. Additionally, between phase II and phase III, there were several notable differences (Bewtra and Lichtenstein Citation2020). Patients in the phase-II study were enrolled from select Italian centers, and therefore, there is a risk that those results might not translate into favorable outcomes in phase III by enrolling a more heterogeneous population of patients from 34 countries. Moreover, the discrepancy in study results between the two trials, despite their similar primary endpoints, could be due to distinct inclusion criteria. Ultimately, these disappointing results serve to emphasize the importance of careful trial design and nuance the perspective of SMAD7 as a drug target in IBD.
Conclusion and perspectives
TGF-β signaling is involved in cell proliferation, differentiation, survival and death across all metazoans. SMAD7, an I-SMAD, negatively regulates TGF-β and BMP signaling through various mechanisms. Similar to R- and Co-SMADs, SMAD7 contains a conserved MH2 domain, enabling type I receptor and R-SMAD interaction. Conversely, its N-terminal region shares limited similarity with R- and Co-SMADs. Of note, the SMAD7 PY motif is critically important to mediate type-I receptor degradation. Overall, it may be said that the structure of SMAD7 underpins its functional divergence, and allows for efficient inhibition of TGF-β and BMP signaling. SMAD7 is regulated at the transcriptional, post-transcriptional and post-translational level, and is subject to a number of PTMs. As the functional relevance of these PTMs is often unclear, this provides avenues for future research. Recently, there has been a marked increase in research addressing the regulation of SMAD7 by ncRNAs. However, how these ncRNAs regulate SMAD7 expression, whether through direct interaction or regulation of an intermediary protein often remains unknown. In addition to its role in TGF-β and BMP signaling, SMAD7 is an important integrator and crosstalk mediator between TGF-β signaling and other signaling pathways, such as MAPK signaling. Since SMAD7 can affect many cellular processes, both by antagonizing TGF-β signaling and by facilitating crosstalk with other signaling pathways, it may be interesting, for instance, to investigate the SMAD7 interactome and PTMs in different cell types. This could lead to a more in depth understanding of how such signaling diversity can be accomplished by SMAD7. This inhibitory SMAD plays an integral role in health and disease and is, among others, important in cancer, fibrosis and IBD. Nonetheless, results from recent clinical trials have painted a more nuanced view of the future of SMAD7 targeting AON in CD.
| Abbreviations | ||
| AhR | = | Aryl hydrocarbon receptor |
| ALK | = | Activin-receptor-like kinase |
| AON | = | Antisense oligonucleotide |
| BMDC | = | Bone marrow-derived cell |
| BMP | = | Bone morphogenetic proteins |
| BMSC | = | Bone marrow mesenchymal stem cell |
| CD | = | Crohn’s disease |
| CIA | = | Collagen-induced arthritis |
| CircRNA | = | Circular RNA |
| CRC | = | Colorectal cancer |
| DEN | = | N-nitrosodiethylamine |
| DSS | = | Dextran sulfate sodium |
| EAE | = | Experimental autoimmune encephalomyelitis |
| ECM | = | Extracellular matrix |
| EGF | = | Epidermal growth factor |
| EMT | = | Epithelial-to-mesenchymal transition |
| EV | = | Extracellular vesicle |
| GBM | = | Glioblastoma |
| GDF | = | Growth and differentiation factor |
| HCC | = | Hepatocellular carcinoma |
| HDAC | = | Histone deacetylases |
| HSC | = | Hepatic stellate cell |
| IBD | = | Inflammatory bowel disease |
| IFN | = | Interferon |
| IL | = | Interleukin |
| LncRNA | = | Long non-coding RNA |
| LRM | = | Leucine rich motif |
| MAPK | = | Mitogen-activated protein kinase |
| MD | = | Molecular dynamics |
| MH | = | MAD homology |
| MIS | = | Müllerian-inhibiting substance |
| MiRNA | = | MicroRNA |
| NcRNA | = | Non-coding RNA |
| OncomiR | = | Oncogenic miRNA |
| Pre-miRNA | = | Precursor miRNA |
| Pri-miRNA | = | Primary miRNA |
| PTM | = | Post-translational modification |
| RA | = | Rheumatoid arthritis |
| RTK | = | Receptor tyrosine kinase |
| SBE | = | SMAD binding element |
| SCC | = | Squamous cell carcinoma |
| SCLC | = | Small cell lung cancer |
| STAT | = | Signal transducer and activator of transcription |
| TAK | = | Transforming growth factor β-activated kinase |
| TGF-β | = | Transforming growth factor β |
| TNF | = | Tumor necrosis factor |
| TPA | = | 12-O-tetradecanoylphorbol-13-acetate |
| TRAF | = | TNF receptor-associated factor |
| UTR | = | Untranslated region |
Disclosure statement
The authors report no declarations of interest.
Additional information
Funding
References
- Abdollah S, Macías-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. 1997. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 272(44):27678–27685.
- Afrakhte M, Morén A, Jossan S, Itoh S, Sampath K, Westermark B, Heldin CH, Heldin NE, Ten Dijke P. 1998. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-β family members . Biochem Biophys Res Commun. 249(2):505–511.
- Akhurst RJ, Hata A. 2012. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 11(10):790–811.
- Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen A-K, Bekker-Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, et al. 2018. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 25(7):631–640. [accessed Jun 14]..
- Alliston T, Ko TC, Cao Y, Liang YY, Feng XH, Chang C, Derynck R. 2005. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem. 280(25):24227–24237.
- Al-Salihi MA, Herhaus L, MacArtney T, Sapkota GP. 2012. USP11 augments TGFβ signaling by deubiquitylating ALK5. Open Biol. 2(6):120063.
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431(7006):350–355.
- Aragón E, Goerner N, Xi Q, Gomes T, Gao S, Massagué J, MacIas MJ. 2012. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-β pathways. Structure. 20(10):1726–1736.
- Ardizzone S, Bevivino G, Monteleone G. 2016. Mongersen, an oral Smad7 antisense oligonucleotide, in patients with active Crohn's disease. Therap Adv Gastroenterol. 9(4):527–532.
- Ayub SG, Kaul D. 2017. miR-2909 regulates ISGylation system via STAT1 signalling through negative regulation of SOCS3 in prostate cancer. Andrology. 5(4):790–797.
- Azuma H, Ehata S, Miyazaki H, Watabe T, Maruyama O, Imamura T, Sakamoto T, Kiyama S, Kiyama Y, Ubai T, et al. 2005. Effect of Smad7 expression on metastasis of mouse mammary carcinoma JygMC(A) cells. J Natl Cancer Inst. 97(23):1734–1746
- Bartel DP. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116(2):281–297.
- Batlle E, Massagué J. 2019. Transforming growth factor-β signaling in immunity and cancer. Immunity. 50(4):924–940.
- Bewtra M, Lichtenstein GR. 2020. Mongersen and SMAD-7 Inhibition, Not a Lucky 7 for Patients With IBD. Am J Gastroenterol. :1.
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Böttinger EP. 2000. A mechanism of suppression of TGF-β/SMAD signaling by NF-kappa B/RelA. Genes Dev. 14(2):187–197.
- Boirivant M, Pallone F, Di Giacinto C, Fina D, Monteleone I, Marinaro M, Caruso R, Colantoni A, Palmieri G, Sanchez M, et al. 2006. Inhibition of Smad7 with a specific antisense oligonucleotide facilitates TGF-β1-mediated suppression of colitis. Gastroenterology. 131(6):1786–1798.
- Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, CORGI Consortium, et al. 2007. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 39(11):1315–1317.
- Brodin G, Åhgren A, Ten Dijke P, Heldin CH, Heuchel R. 2000. Efficient TGF-β induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter . J Biol Chem. 275(37):29023–29030.
- Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Varanko AK, Xu Y, et al. 2015. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat Commun. 6(1).
- Casado P, Alcolea MP, Iorio F, Rodríguez-Prados JC, Vanhaesebroeck B, Saez-Rodriguez J, Joel S, Cutillas PR. 2013. Phosphoproteomics data classify hematological cancer cell lines according to tumor type and sensitivity to kinase inhibitors. Genome Biol. 14(4):R37.
- Chang CH, Yen MC, Liao SH, Hsu YL, Lai CS, Kuo YR, Hsu YL. 2017. Dual role of MiR-21-mediated signaling in HUVECs and rat surgical flap under normoxia and hypoxia condition. Int J Mol Sci. 18(9)
- Chang L, Yuan Y, Li C, Guo T, Qi H, Xiao Y, Dong X, Liu Z, Liu Q. 2016. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 383(2):183–194.
- Chang Y, Liu C, Yang J, Liu G, Feng F, Tang J, Hu L, Li L, Jiang F, Chen C, et al. 2013. MiR-20a triggers metastasis of gallbladder carcinoma. J Hepatol. 59(3):518–527.
- Chao LI, Hua-Yu ZHU, Wen-Dong BAI, Mei S, Bin X, Da-Hai HU, Yi LIU. 2019. miR-96 promotes collagen deposition in keloids by targeting Smad7. Exp Ther Med. 17(1):773–781.
- Che H, Wang Y, Li Y, Lv J, Li H, Liu Y, Dong R, Sun Y, Xu X, Zhao J, et al. 2020. Inhibition of microRNA-150-5p alleviates cardiac inflammation and fibrosis via targeting Smad7 in high glucose-treated cardiac fibroblasts. J Cell Physiol. 235(11):7769–7779.
- Chen B, Chen X, Wu X, Wang X, Wang Y, Lin TY, Kurata J, Wu J, Vonderfecht S, Sun G, et al. 2015. Disruption of microRNA-21 by TALEN leads to diminished cell transformation and increased expression of cell-environment interaction genes. Cancer Lett. 356(2 Pt B):506–516.
- Chen E, Li Q, Wang H, Yang F, Min L, Yang J. 2018. MiR-92a promotes tumorigenesis of colorectal cancer, a transcriptomic and functional based study. Biomed Pharmacother. 106:1370–1377.
- Chen G, Cao S, Liu F, Liu Y. 2015. miR-195 plays a role in steroid resistance of ulcerative colitis by targeting Smad7. Biochem J. 471(3):357–367.
- Chen G, Huang S, Song F, Zhou Y, He X. 2020. Lnc-Ang362 is a pro-fibrotic long non-coding RNA promoting cardiac fibrosis after myocardial infarction by suppressing Smad7. Arch Biochem Biophys. 685:108354.
- Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M, Teal CB, Brem RF, Stojadinovic A, Grinkemeyer M, et al. 2013. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS One. 8(1):e54213.
- Chen Y, Sun C, Lu J, Zou L, Hu M, Yang Z, Xu Y. 2019. MicroRNA-590-5p antagonizes the inhibitory effect of high glucose on osteoblast differentiation by suppressing Smad7 in MC3T3-E1 cells. J Int Med Res. 47(4):1740–1748.
- Chen Y, Zhang Q, Zhou Y, Yang Z, Tan M. 2020. Inhibition of miR-182-5p attenuates pulmonary fibrosis via TGF-β/Smad pathway. Hum Exp Toxicol. 39(5):683–695.
- Chong PA, Lin H, Wrana JL, Forman-Kay JD. 2006. An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J Biol Chem. 281(25):17069–17075.
- Chung ACK, Huang XR, Zhou L, Heuchel R, Lai KN, Lan HY. 2009. Disruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in mice. Nephrol Dial Transplant. 24(5):1443–1454.
- Clevers H, Nusse R. 2012. Wnt/β-catenin signaling and disease. Cell. 149(6):1192–1205.
- Cutts AJ, Soond SM, Powell S, Chantry A. 2011. Early phase TGFβ receptor signalling dynamics stabilised by the deubiquitinase UCH37 promotes cell migratory responses. Int J Biochem Cell Biol. 43(4):604–612.
- Dai F, Liu T, Zheng S, Liu Q, Yang C, Zhou J, Chen Y, Sheyhidin I, Lu X. 2016. MiR-106b promotes migration and invasion through enhancing EMT via downregulation of Smad 7 in Kazakh's esophageal squamous cell carcinoma. Tumour Biol. 37(11):14595–14604.
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. 2010. Smad proteins bind a conserved RNA sequence to promote MicroRNA maturation by Drosha. Mol Cell. 39(3):373–384.
- De Simone V, Bevivino G, Sedda S, Izzo R, Laudisi F, Dinallo V, Franzè E, Colantoni A, Ortenzi A, Salvatori S, et al. 2017. Smad7 knockdown activates protein kinase RNA-associated eIF2α pathway leading to colon cancer cell death. Cell Death Dis. 8(3):e2681–e2681.
- Derynck R, Budi EH. 2019. Specificity, versatility, and control of TGF-b family signaling. Sci Signal. 12(570):eaav5183.
- Derynck R, Zhang Y, Feng XH. 1998. Smads: Transcriptional activators of TGF-β responses. Cell. 95(6):737–740.
- Dooley S, Hamzavi J, Ciuclan L, Godoy P, Ilkavets I, Ehnert S, Ueberham E, Gebhardt R, Kanzler S, Geier A, et al. 2008. Hepatocyte-Specific Smad7 Expression Attenuates TGF-β-Mediated Fibrogenesis and Protects Against Liver Damage. Gastroenterology. 135(2):642–659.e46.
- Dooley S, Said HM, Gressner AM, Floege J, En-Nia A, Mertens PR. 2006. Y-box protein-1 is the crucial mediator of antifibrotic interferon-gamma effects. J Biol Chem. 281(3):1784–1795.
- Dowdy SC, Mariani A, Janknecht R. 2003. HER2/Neu- and TAK1-mediated Up-regulation of the Transforming Growth Factor β inhibitor Smad7 via the ETS protein ER81. J Biol Chem. 278(45):44377–44384.
- Du J, Zheng R, Xiao F, Zhang S, He K, Zhang J, Shao Y. 2017. Downregulated microRNA-195 in the bicuspid aortic valve promotes calcification of valve interstitial cells via targeting SMAD7. Cell Physiol Biochem. 44(3):884–896.
- Duan Y, Chen Q. 2016. TGF-β1 regulating MIR-205/MIR-195 expression affects the TGF-β signal pathway by respectively targeting SMAD2/SMAD7. Oncol Rep. 36(4):1837–1844.
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. 2001. Smurf1 Interacts with Transforming Growth factor-β type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 276(16):12477–12480.
- Edlund S, Bu S, Schuster N, Aspenström P, Heuchel R, Heldin NE, Ten Dijke P, Heldin CH, Landström M. 2003. Transforming growth factor-β1 (TGF-β)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-β-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell. 14(2):529–544.
- Edlund S, Lee SY, Grimsby S, Zhang S, Aspenström P, Heldin C-H, Landström M. 2005. Interaction between Smad7 and β-catenin: importance for transforming growth factor β-induced apoptosis. Mol Cell Biol. 25(4):1475–1488.
- Eichhorn PJA, Rodón L, Gonzàlez-Juncà A, Dirac A, Gili M, Martínez-Sáez E, Aura C, Barba I, Peg V, Prat A, et al. 2012. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med. 18(3):429–435.
- Elkouris M, Kontaki H, Stavropoulos A, Antonoglou A, Nikolaou KC, Samiotaki M, Szantai E, Saviolaki D, Brown PJ, Sideras P, et al. 2016. SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep. 15(12):2733–2744.
- Ezzie ME, Crawford M, Cho JH, Orellana R, Zhang S, Gelinas R, Batte K, Yu L, Nuovo G, Galas D, et al. 2012. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 67(2):122–131.
- Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA Translation and Stability by microRNAs. Annu Rev Biochem. 79(1):351–379.
- Fang SH, Chen L, Chen HH, Li YF, Luo HB, Hu DQ, Chen P. 2019. MiR-15b ameliorates SONFH by targeting Smad7 and inhibiting osteogenic differentiation of BMSCs. Eur Rev Med Pharmacol Sci. 23(22):9761–9771.
- Fantini MC, Rizzo A, Fina D, Caruso R, Sarra M, Stolfi C, Becker C, MacDonald TT, Pallone F, Neurath MF, et al. 2009. Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology. 136(4):1308–1316.e3.
- Feagan BG, Sands BE, Rossiter G, Li X, Usiskin K, Zhan X, Colombel JF. 2018. Effects of Mongersen (GED-0301) on endoscopic and clinical outcomes in patients with active Crohn’s disease. Gastroenterology. 154(1):61–64.e6.
- Feng M, Tang PMK, Huang XR, Sun SF, You YK, Xiao J, Lv LL, Xu AP, Lan HY. 2018. TGF-β mediates renal fibrosis via the Smad3-Erbb4-IR long noncoding RNA axis. Mol Ther. 26(1):148–161.
- Feng T, Dzieran J, Yuan X, Dropmann A, Maass T, Teufel A, Marhenke S, Gaiser T, Rückert F, Kleiter I, et al. 2017. Hepatocyte-specific Smad7 deletion accelerates DEN-induced HCC via activation of STAT3 signaling in mice. Oncogenesis. 6(1):e294
- Feng YH, Tsao CJ. 2016. Emerging role of microRNA-21 in cancer. Biomed Rep. 5(4):395–402.
- Ferrigno O, Lallemand F, Verrecchia F, L'Hoste S, Camonis J, Atfi A, Mauviel A. 2002. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-β/Smad signaling. Oncogene. 21(32):4879–4884.
- Fortini BK, Tring S, Plummer SJ, Edlund CK, Moreno V, Bresalier RS, Barry EL, Church TR, Figueiredo JC, Casey G. 2014. Multiple functional risk variants in a SMAD7 enhancer implicate a colorectal cancer risk haplotype. PLoS One. 9(11):e111914.
- Fu Y, Tang Y, Wang J, Guo Z. 2019. MicroRNA-181c suppresses the biological progression of osteosarcoma via targeting smad7 and regulating transforming growth factor-β (TGF-β) signaling pathway. Med Sci Monit. 25:4801–4810.
- Gao X, Xu H, Xu D, Li S, Wei Z, Li S, Cai W, Mao N, Jin F, Li Y. 2020. MiR-411–3p alleviates Silica-induced pulmonary fibrosis by regulating Smurf2/TGF-β signaling. Exp Cell Res. 388(2).
- Garo LP, Ajay AK, Fujiwara M, Beynon V, Kuhn C, Gabriely G, Sadhukan S, Raheja R, Rubino S, Weiner HL, et al. 2019. Smad7 controls immunoregulatory PDL2/1-PD1 signaling in intestinal inflammation and autoimmunity. Cell Rep. 28(13):3353–3366.e5.
- Garros RF, Paul R, Connolly M, Lewis A, Garfield BE, Natanek SA, Bloch S, Mouly V, Griffiths MJ, Polkey MI, et al. 2017. MicroRNA-542 promotes mitochondrial dysfunction and SMAD activity and is elevated in intensive care unit-acquired Weakness. Am J Respir Crit Care Med. 196(11):1422–1433.
- Ghorbani S, Talebi F, Chan WF, Masoumi F, Vojgani M, Power C, Noorbakhsh F. 2017. MicroRNA-181 variants regulate T cell phenotype in the context of autoimmune neuroinflammation. Front Immunol. 8:758.
- Gohla G, Krieglstein K, Spittau B. 2008. Tieg3/Klf11 induces apoptosis in OLI-neu cells and enhances the TGF-β signaling pathway by transcriptional repression of Smad7. J Cell Biochem. 104(3):850–861.
- Grau AM, Datta PK, Zi J, Halder SK, Beauchamp RD. 2006. Role of Smad proteins in the regulation of NF-kappaB by TGF-β in colon cancer cells. Cell Signal. 18(7):1041–1050.
- Grönroos E, Hellman U, Heldin CH, Ericsson J. 2002. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 10(3):483–493.
- Gu W, Hong X, Le Bras A, Nowak WN, Bhaloo SI, Deng J, Xie Y, Hu Y, Ruan XZ, Xu Q. 2018. Smooth muscle cells differentiated from mesenchymal stem cells are regulated by microRNAs and suitable for vascular tissue grafts. J Biol Chem. 293(21):8089–8102.
- Ha Thi HT, Kim H-Y, Choi S-W, Kang J-M, Kim S-J, Hong S. 2015. Smad7 Modulates Epidermal Growth Factor Receptor Turnover through Sequestration of c-Cbl. Mol Cell Biol. 35(16):2841–2850.
- Ha Thi HT, Kim H-Y, Lee Y-J, Kim S-J, Hong S. 2019. SMAD7 in keratinocytes promotes skin carcinogenesis by activating ATM-dependent DNA repair and an EGFR-mediated cell proliferation pathway | Carcinogenesis | Oxford Academic. Carcinogenesis. 40(1):112–120.
- Halder SK, Beauchamp RD, Datta PK. 2005. Smad7 induces tumorigenicity by blocking TGF-β-induced growth inhibition and apoptosis. Exp Cell Res. 307(1):231–246.
- Halder SK, Rachakonda G, Deane NG, Datta PK. 2008. Smad7 induces hepatic metastasis in colorectal cancer. Br J Cancer. 99(6):957–965.
- Hamzavi J, Ehnert S, Godoy P, Ciuclan L, Weng H, Mertens PR, Heuchel R, Dooley S. 2008. Disruption of the Smad7 gene enhances CCI4-dependent liver damage and fibrogenesis in mice. J Cell Mol Med. 12(5B):2130–2144.
- Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, ten Dijke P, Lin X, et al. 2006. Smad7-Induced β-catenin degradation alters epidermal appendage development. Dev Cell. 11(3):301–312.
- Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. 2001. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-β signaling. J Cell Biol. 155(6):1017–1027.
- Hariharan R, Pillai MR. 2008. Structure-function relationship of inhibitory Smads: Structural flexibility contributes to functional divergence. Proteins Struct Funct Genet. 71(4):1853–1862.
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Wrana JL, et al. 1997. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 89(7):1165–1173.
- He G, Chen J, Huang D. 2019. miR-877-3p promotes TGF-β1-induced osteoblast differentiation of MC3T3-E1 cells by targeting Smad7. Exp Ther Med. 18(1):312–319
- He J, Sun X, Qian KQ, Liu X, Wang Z, Chen Y. 2009. Protection of cerulein-induced pancreatic fibrosis by pancreas-specific expression of Smad7. Biochim Biophys Acta - Mol Basis Dis. 1792(1):56–60.
- Heldin CH, Moustakas A. 2016. Signaling receptors for TGF-β family members. Cold Spring Harb Perspect Biol. 8(8):a022053.
- Herhaus L, Sapkota GP. 2014. The emerging roles of deubiquitylating enzymes (DUBs) in the TGFβ and BMP pathways. Cell Signal. 26(10):2186–2192.
- Hill CS. 2016. Transcriptional control by the SMADs. Cold Spring Harb Perspect Biol. 8(10):a022079.
- Hong S, Lim S, Li AG, Lee C, Lee YS, Lee EK, Park SH, Wang XJ, Kim SJ. 2007. Smad7 binds to the adaptors TAB2 and TAB3 to block recruitment of the kinase TAK1 to the adaptor TRAF2. Nat Immunol. 8(5):504–513.
- Hsu YL, Huang MS, Hung JY, Chang WA, Tsai YM, Pan YC, Lin YS, Tsai HP, Kuo PL. 2020. Bone-marrow-derived cell-released extracellular vesicle miR-92a regulates hepatic pre-metastatic niche in lung cancer. Oncogene. 39(4):739–753.
- Hu B, Mao Z, Du Q, Jiang X, Wang Z, Xiao Z, Zhu D, Wang X, Zhu Y, Wang H. 2019. miR-93-5p targets Smad7 to regulate the transforming growth factor-β1/Smad3 pathway and mediate fibrosis in drug-resistant prolactinoma. Brain Res Bull. 149:21–31.
- Hu HH, Chen DQ, Wang YN, Feng YL, Cao G, Vaziri ND, Zhao YY. 2018. New insights into TGF-β/Smad signaling in tissue fibrosis. Chem Biol Interact. 292:76–83.
- Hu J, Xu J-F, Ge W-L. 2016. MiR-497 enhances metastasis of oral squamous cell carcinoma through SMAD7 suppression. Am J Transl Res. 8(7):3023–3031.
- Hua X, Miller ZA, Benchabane H, Wrana JL, Lodish HF. 2000. Synergism between transcription factors TFE3 and Smad3 in transforming growth factor-β-induced transcription of the Smad7 gene. J Biol Chem. 275(43):33205–33208.
- Huang C, Song H, Lai L. 2019. The role and mechanism of microRNA-18a-5p in oral squamous cell carcinoma. Mol Med Rep. 20(2):1637–1644.
- Huang CF, Sun CC, Zhao F, Zhang YD, Li DJ. 2015. miR-33a levels in hepatic and serum after chronic HBV-induced fibrosis. J Gastroenterol. 50(4):480–490.
- Huang N, Li W, Wang X, Qi S. 2018. MicroRNA-17-5p aggravates lipopolysaccharide-induced injury in nasal epithelial cells by targeting Smad7. BMC Cell Biol. 19(1):1
- Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, Janitz M. 2017. The emerging role of circular RNAs in transcriptome regulation. Genomics. 109(5-6):401–407.
- Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, Huang X, Liang S, Chen R, Chen S, et al. 2020. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 27(5):1709–1727.
- Hujie G, Zhou S. H, Zhang H, Qu J, Xiong X, Wei Hujie O, Liao C. G, Yang S. E. 2018. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/KLF11/Smads in hepatocellular carcinoma. Cancer Cell Int. 18(1):10.
- Ibarrola N, Kratchmarova I, Nakajima D, Schiemann WP, Moustakas A, Pandey A, Mann M. 2004. Cloning of a novel signaling molecule, AMSH-2, that potentiates transforming growth factor β signaling. BMC Cell Biol. 5:2.
- Ihara S, Hirata Y, Koike K. 2017. TGF-β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J Gastroenterol. 52(7):777–787.
- Imamura T, Oshima Y, Hikita A. 2013. Regulation of TGF-β family signalling by ubiquitination and deubiquitination. J Biochem. 154(6):481–489.
- Inamitsu M, Itoh S, Hellman U, ten Dijke P, Kato M. 2006. Methylation of Smad6 by protein arginine N-methyltransferase 1. FEBS Lett. 580(28-29):6603–6611.
- Itoh S, Landström M, Hermansson A, Itoh F, Heldin CH, Heldin NE, Ten Dijke P. 1998. Transforming growth factor β1 induces nuclear export of inhibitory Smad7. J Biol Chem. 273(44):29195–29201.
- Itoh S, ten Dijke P. 2007. Negative regulation of TGF-β receptor/Smad signal transduction. Curr Opin Cell Biol. 19(2):176–184.
- Jairath V, Zou G, Parker CE, MacDonald JK, Mosli MH, AlAmeel T, Al Beshir M, AlMadi M, Al-Taweel T, Atkinson NSS, et al. 2017. Systematic review with meta-analysis: placebo rates in induction and maintenance trials of Crohn's disease. Aliment Pharmacol Ther. 45(8):1021–1042.
- Javelaud D, Delmas V, Möller M, Sextius P, André J, Menashi S, Larue L, Mauviel A. 2005. Stable overexpression of Smad7 in human melanoma cells inhibits their tumorigenicity in vitro and in vivo. Oncogene. 24(51):7624–7629.
- Javelaud D, Mohammad KS, McKenna CR, Fournier P, Luciani F, Niewolna M, André J, Delmas V, Larue L, Guise TA, et al. 2007. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 67(5):2317–2324.
- Jenkins BJ, Grail D, Nheu T, Najdovska M, Wang B, Waring P, Inglese M, McLoughlin RM, Jones SA, Topley N, et al. 2005. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-β signaling. Nat Med. 11(8):845–852.
- Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu X, Feng Y, Dai Z. 2014. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp Mol Med. 46(7):e107–e107.
- Jiao C, Li L, Zhang P, Zhang L, Li K, Fang R, Yuan L, Shi K, Pan L, Guo Q, et al. 2020a. REGγ ablation impedes dedifferentiation of anaplastic thyroid carcinoma and accentuates radio-therapeutic response by regulating the Smad7-TGF-β pathway. Cell Death Differ. 27(2):497–508.
- Jiao C, Li L, Zhang P, Zhang L, Li K, Fang R, Yuan L, Shi K, Pan L, Guo Q, et al. 2020b. REGγ ablation impedes dedifferentiation of anaplastic thyroid carcinoma and accentuates radio-therapeutic response by regulating the Smad7-TGF-β pathway. Cell Death Differ. 27(2):497–508.
- Jiao H, Xie D, Qiao Y. 2019. LncRNA PRINS is involved in the development of nephropathy in patients with diabetes via interaction with Smad7. Exp Ther Med. 17(4):3203–3208.
- Jungert K, Buck A, Buchholz M, Wagner M, Adler G, Gress TM, Ellenrieder V. 2006. Smad-Sp1 complexes mediate TGFβ-induced early transcription of oncogenic Smad7 in pancreatic cancer cells. Carcinogenesis. 27(12):2392–2401.
- Kamato D, Burch ML, Piva TJ, Rezaei HB, Rostam MA, Xu S, Zheng W, Little PJ, Osman N. 2013. Transforming growth factor-β signalling: Role and consequences of Smad linker region phosphorylation. Cell Signal. 25(10):2017–2024.
- Kan H, Guo W, Huang Y, Liu D. 2015. MicroRNA-520g induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by targeting SMAD7. FEBS Lett. 589(1):102–109.
- Katsuno Y, Qin J, Oses-Prieto J, Wang H, Jackson-Weaver O, Zhang T, Lamouille S, Wu J, Burlingame A, Xu J, et al. 2018. Arginine methylation of SMAD7 by PRMT1 in TGF-–induced epithelial–mesenchymal transition and epithelial stem-cell generation. J Biol Chem. 293(34):13059–137072.
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol Cell. 6(6):1365–1375.
- Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 10(2):126–139.
- Kit Leng Lui S, Iyengar PV, Jaynes P, Isa ZFBA, Pang B, Tan TZ, Eichhorn PJA. 2017. USP26 regulates TGF-β signaling by deubiquitinating and stabilizing SMAD7. EMBO Rep. 18(5):797–808.
- Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura T, et al. 2003. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. Embo J. 22(24):6458–6470.
- Kostopoulou F, Malizos KN, Papathanasiou I, Tsezou A. 2015. MicroRNA-33a regulates cholesterol synthesis and cholesterol efflux-related genes in osteoarthritic chondrocytes. Arthritis Res Ther. 17(1):42
- Kruse JJCM, Floot BGJ, Te Poele JAM, Russell NS, Stewart FA. 2009. Radiation-Induced Activation of TGF-β signaling pathways in relation to vascular damage in mouse kidneys. Radiat Res. 171(2):188–197.
- Kuang C, Xiao Y, Liu X, Stringfield TM, Zhang S, Wang Z, Chen Y. 2006. In vivo disruption of TGF-β signaling by Smad7 leads to premalignant ductal lesions in the pancreas. Proc Natl Acad Sci USA. 103(6):1858–1863.
- Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T. 2005. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor. Biochem J. 386(Pt 3):461–470.
- Lallemand F, Seo SR, Ferrand N, Pessah M, L'Hoste S, Rawadi G, Roman-Roman S, Camonis J, Atfi A. 2005. AIP4 restricts transforming growth factor-β signaling through a ubiquitination-independent mechanism. J Biol Chem. 280(30):27645–27653.
- Lamora A, Talbot J, Bougras G, Amiaud J, Leduc M, Chesneau J, Taurelle J, Stresing V, Le Deley MC, Heymann MF, et al. 2014. Overexpression of Smad7 blocks primary tumor growth and lung metastasis development in osteosarcoma. Clin Cancer Res. 20(19):5097–5112.
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129(7):1401–1414.
- Leivonen SK, Ala-Aho R, Koli K, Grénman R, Peltonen J, Kähäri VM. 2006. Activation of Smad signaling enhances collagenase-3 (MMP-13) expression and invasion of head and neck squamous carcinoma cells. Oncogene. 25(18):2588–2600.
- Li H, Li J, Chen L, Qi S, Yu S, Weng Z, Hu Z, Zhou Q, Xin Z, Shi L, et al. 2019. HERC3-Mediated SMAD7 ubiquitination degradation promotes autophagy-induced EMT and chemoresistance in Glioblastoma. Clin Cancer Res. 25(12):3602–3616.
- Li PF, He RH, Shi SB, Li R, Wang QT, Rao GT, Yang B. 2019. Modulation of miR-10a-mediated TGF-β1/Smads signaling affects atrial fibrillation-induced cardiac fibrosis and cardiac fibroblast proliferation. Biosci Rep. 39(2):BSR20181931.
- Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, Wang W, Zhang L, Zhang X, Tang Q, et al. 2013. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 335(1):168–174.
- Li S, Fan Q, He S, Tang T, Liao Y, Xie J. 2015. MicroRNA-21 negatively regulates treg cells through a TGF-β1/smad-independent pathway in patients with coronary heart disease. Cell Physiol Biochem. 37(3):866–878.
- Li X, Guo L, Liu Y, Su Y, Xie Y, Du J, Wang S, Wang H, Liu Y. 2018. MicroRNA-21 promotes wound healing via the Smad7-Smad2/3-Elastin pathway. Exp Cell Res. 362(2):245–251.
- Li X, Nania S, Fejzibegovic N, Moro CF, Klopp-Schulze L, Verbeke C, Löhr JM, Heuchel RL. 2016. Cerulein-induced pancreatic fibrosis is modulated by Smad7, the major negative regulator of transforming growth factor-β signaling. Biochim Biophys Acta - Mol Basis Dis. 1862(9):1839–1846.
- Li X, Zeng X. 2020. Shikonin suppresses progression and epithelial-mesenchymal transition in hepatocellular carcinoma (HCC) cells by modulating miR-106b/SMAD7/TGF-β signaling pathway. Cell Biol Int. 44(2):467–476.
- Li Y, Wang H, Li J, Yue W. 2014. MiR-181c modulates the proliferation, migration, and invasion of neuroblastoma cells by targeting Smad7. Acta Biochim Biophys Sin (Shanghai)). 46(1):48–55.
- Li Y, Xiang Y, Song Y, Wan L, Yu G, Tan L. 2019. Dysregulated miR-142, -33b and -423 in granulosa cells target TGFBR1 and SMAD7: a possible role in polycystic ovary syndrome. Mol Hum Reprod. 25(10):638–646.
- Liang ZG, Yao H, Xie RS, Gong CL, Tian Y. 2018. MicroRNA-20b-5p promotes ventricular remodeling by targeting the TGF-β/Smad signaling pathway in a rat model of ischemia-reperfusion injury. Int J Mol Med. 42(2):975–987.
- Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. 2010. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 207(8):1589–1597.
- Liu H, Li S, Jiang W, Li Y. 2020. MiR-484 protects rat myocardial cells from ischemia-reperfusion injury by inhibiting caspase-3 and caspase-9 during apoptosis. Korean Circ J. 50(3):250–263.
- Liu J, Zhou Y, Shi Z, Hu Y, Meng T, Zhang X, Zhang S, Zhang J. 2016. MicroRNA-497 Modulates Breast Cancer Cell Proliferation, Invasion, and Survival by Targeting SMAD7. DNA Cell Biol. 35(9):521–529.
- Liu N, Jiao T, Huang Y, Liu W, Li Z, Ye X. 2015. Hepatitis B Virus Regulates Apoptosis and Tumorigenesis through the MicroRNA-15a-Smad7-Transforming Growth Factor β Pathway. J Virol. 89(5):2739–2749.
- Liu T, Wu Y, Huang T, Zhang X, Cai Y. 2017. miR-590 promotes the proliferation of HUMSCs and induces ECM synthesis by targeting Smad7. Oncol Lett. 14(4):3941–3946.
- Liu W, Rui H, Wang J, Lin S, He Y, Chen M, Li Q, Ye Z, Zhang S, Siu CC, et al. 2006. Axin is a scaffold protein in TGF-β signaling that promotes degradation of Smad7 by Arkadia. Embo J. 25(8):1646–1658.
- Liu X, Lee J, Cooley M, Bhogte E, Hartley S, Glick A. 2003. Smad7 but not Smad6 Cooperates with Oncogenic ras to Cause Malignant Conversion in a Mouse Model for Squamous Cell Carcinoma. [place unknown].
- Liu Z, Huang XR, Chen HY, Fung E, Liu J, Lan HY. 2017. Deletion of angiotensin-converting enzyme-2 promotes hypertensive nephropathy by targeting smad7 for ubiquitin degradation. Hypertension. 70(4):822–830.
- Li Y, Cui C, Feng X, Kielbasa S, Mei H, Van Dinther M, Van Dam H, Bauer A, Zhang L, Ten Dijke P. 2020. VprBP Mitigates TGF-β and Activin Signaling by Promoting Smurf1-mediated Type I Receptor Degradation. J Mol Cell Biol. 12(2):138–151.
- Luo M, Tan X, Mu L, Luo Y, Li R, Deng X, Chen N, Ren M, Li Y, Wang L, et al. 2017. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci Rep. 7:43427.
- Luo X, Ding Q, Wang M, Li Z, Mao K, Sun B, Pan Y, Wang Z, Zang YQ, Chen Y. 2010. In vivo disruption of TGF-β Signaling by Smad7 in airway epithelium alleviates allergic asthma but aggravates lung carcinogenesis in mouse. PLoS One. 5(4):e10149.
- Luo X, Zhang D, Xie J, Su Q, He X, Bai R, Gao G, Pan W. 2018. MicroRNA-96 Promotes Schistosomiasis Hepatic Fibrosis in Mice by Suppressing Smad7. Mol Ther Methods Clin Dev. 11:73–82.
- Luwor RB, Baradaran B, Taylor LE, Iaria J, Nheu TV, Amiry N, Hovens CM, Wang B, Kaye AH, Zhu HJ. 2013. Targeting Stat3 and Smad7 to restore TGF-β cytostatic regulation of tumor cells in vitro and in vivo. Oncogene. 32(19):2433–2441.
- Lv H, Nan Z, Jiang P, Wang Z, Song M, Ding H, Liu D, Zhao G, Zheng Y, Hu Y. 2019. Vascular endothelial growth factor 165 inhibits pro-fibrotic differentiation of stromal cells via the DLL4/Notch4/smad7 pathway. Cell Death Dis. 10(9):1–17.
- Malonis RJ, Fu W, Jelcic MJ, Thompson M, Canter BS, Tsikitis M, Esteva FJ, Sánchez I. 2017. RNF11 sequestration of the E3 ligase SMURF2 on membranes antagonizes SMAD7 down-regulation of transforming growth factor β signaling. J Biol Chem. 292(18):7435–7451.
- Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, Christensen RN, Schmidt WN, McCaffrey AP. 2010. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 90(12):1727–1736.
- Massagué J. 1998. TGF-β signal transduction. Annu Rev Biochem. 67(1):753–791.
- Massagué J. 2008. TGFβ in Cancer. Cell. 134(2):215–230.
- Massagué J. 2012. TGFβ signalling in context. Nat Rev Mol Cell Biol. 13(10):616–630.
- Massagué J, Wotton D. 2000. Transcriptional Control by the TGF-β/Smad Signaling System. Embo J. 19(8):1745–1754.
- Matsuzaki K. 2011. Smad Phosphoisoform Signaling Specificity: The Right Place at the Right Time. Carcinogenesis. 32(11):1578–1588.
- Mazars A, Lallemand F, Prunier C, Marais J, Ferrand N, Pessah M, Cherqui G, Atfi A. 2001. Evidence for a role of the JNK cascade in Smad7-mediated apoptosis. J Biol Chem. 276(39):36797–36803.
- Meng Z, Moroishi T, Guan KL. 2016. Mechanisms of Hippo pathway regulation. Genes Dev. 30(1):1–17.
- Miyazawa K, Miyazono K. 2017. Regulation of TGF-β family signaling by inhibitory smads. Cold Spring Harb Perspect Biol. 9(3):a022095.
- Mochizuki T, Miyazaki H, Hara T, Furuya T, Imamura T, Watabe T, Miyazono K. 2004. Roles for the MH2 domain of Smad7 in the specific inhibition of transforming growth factor-β superfamily signaling. J Biol Chem. 279(30):31568–31574.
- Monteleone G, Del Vecchio Blanco G, Monteleone I, Fina D, Caruso R, Gioia V, Ballerini S, Federici G, Bernardini S, Pallone F, et al. 2005. Post-transcriptional regulation of Smad7 in the gut of patients with inflammatory bowel disease. Gastroenterology. 129(5):1420–1429.
- Monteleone G, Fantini MC, Onali S, Zorzi F, Sancesario G, Bernardini S, Calabrese E, Viti F, Monteleone I, Biancone L, et al. 2012. Phase I clinical trial of SMAD7 knockdown using antisense oligonucleotide in patients with active Crohn's disease. Mol Ther. 20(4):870–876.
- Monteleone I, Marafini I, Zorzi F, Fusco DD, Dinallo V, Rizzo A, Sileri P, Sica G, Monteleone G. 2016. Smad7 knockdown restores aryl hydrocarbon receptor-mediated protective signals in the gut. J Crohns Colitis. 10(6):670–677.
- Morikawa M, Derynck R, Miyazono K. 2016. TGF- β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 8(5):a021873.
- Mota MS, Jackson WP, Bailey SK, Vayalil P, Landar A, Rostas JW, Mulekar MS, Samant RS, Shevde LA. 2018. Deficiency of tumor suppressor merlin facilitates metabolic adaptation by co-operative engagement of SMAD-Hippo signaling in breast cancer. Carcinogenesis. 39(9):1165–1175.
- Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. 2003. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 14(7):2809–2817.
- Murugaiyan G, Da Cunha AP, Ajay AK, Joller N, Garo LP, Kumaradevan S, Yosef N, Vaidya VS, Weiner HL. 2015. MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Invest. 125(3):1069–1080.
- Nagarajan RP, Chen F, Li W, Vig E, Harrington MA, Nakshatri H, Chen Y. 2000. Repression of transforming-growth-factor-β-mediated transcription by nuclear factor κB. Biochem J. 348(3):591–596.
- Nagarajan RP, Zhang J, Li W, Chen Y. 1999. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J Biol Chem. 274(47):33412–33418.
- Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, et al. 1997. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature. 389(6651):631–635.
- Nakao A, Fujii M, Matsumura R, Kumano K, Saito Y, Miyazono K, Iwamoto I. 1999. Transient gene transfer and expression of Smad7 prevents bleomycin-induced lung fibrosis in mice. J Clin Invest. 104(1):5–11.
- Napetschnig J, Wu H. 2013. Molecular basis of NF-κB signaling. Annu Rev Biophys. 42(1):443–468.
- Nong Q, Li S, Wu Y, Liu D. 2018. LncRNA COL1A2-AS1 inhibits the scar fibroblasts proliferation via regulating miR-21/Smad7 pathway. Biochem Biophys Res Commun. 495(1):319–324.
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. 2005. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 19(3):297–308.
- Parikh A, Lee C, Joseph P, Marchini S, Baccarini A, Kolev V, Romualdi C, Fruscio R, Shah H, Wang F, et al. 2014. MicroRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat Commun. 5(1):1–16.
- Park S, Kang JM, Kim SJ, Kim H, Hong S, Lee YJ, Kim S-J. 2015. Smad7 enhances ATM activity by facilitating the interaction between ATM and Mre11-Rad50-Nbs1 complex in DNA double-strand break repair. Cell Mol Life Sci. 72(3):583–596.
- Park SH, Jung EH, Kim GY, Kim BC, Lim JH, Woo CH. 2015. Itch E3 Ubiquitin Ligase Positively Regulates TGF-β Signaling to EMT via Smad7 Ubiquitination. Mol Cells. 38(1):20–25.
- Patil S, Wildey GM, Brown TL, Choy L, Derynck R, Howe PH. 2000. Smad7 is induced by CD40 and protects WEHI 231 B-lymphocytes from transforming growth factor-β -induced growth inhibition and apoptosis. J Biol Chem. 275(49):38363–38370.
- Pearson G, Robinson F, Beers Gibson T, Xu B, Karandikar M, Berman K, Cobb MH. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions*. Endocr Rev. 22(2):153–183.
- Pulaski L, Landström M, Heldin CH, Souchelnytskyi S. 2001. Phosphorylation of Smad7 at Ser-249 does not interfere with its inhibitory role in transforming growth factor-β-dependent signaling but affects Smad7-dependent transcriptional activation. J Biol Chem. 276(17):14344–14349.
- Pulignani S, Borghini A, Foffa I, Vecoli C, Ait-Alì L, Andreassi MG. 2020. Functional characterization and circulating expression profile of dysregulated microRNAs in BAV-associated aortopathy. Heart Vessels. 35(3):432–440.
- Qin Z, Xia W, Fisher GJ, Voorhees JJ, Quan T. 2018. YAP/TAZ regulates TGF-β/Smad3 signaling by induction of Smad7 via AP-1 in human skin dermal fibroblasts. Cell Commun Signal. 16(1):18
- Quan T, He T, Voorhees JJ, Fisher GJ. 2001. Ultraviolet Irradiation Blocks Cellular Responses to Transforming Growth factor-β by down-regulating its type-II receptor and inducing Smad7. J Biol Chem. 276(28):26349–26356.
- Ratz L, Laible M, Kacprzyk LA, Wittig-Blaich SM, Tolstov Y, Duensing S, Altevogt P, Klauck SM, Sültmann H. 2017. TMPRSS2:ERG gene fusion variants induce TGF-β signaling and epithelial to mesenchymal transition in human prostate cancer cells. Oncotarget. 8(15):25115–25130.
- Rawlings JS, Rosler KM, Harrison DA. 2004. The JAK/STAT signaling pathway. J Cell Sci. 117(Pt 8):1281–1283.
- Rizzo A, De Mare V, Rocchi C, Stolfi C, Colantoni A, Neurath MF, Macdonald TT, Pallone F, Monteleone G, Fantini MC. 2014. Smad7 induces plasticity in tumor-infiltrating Th17 cells and enables TNF-alpha-mediated killing of colorectal cancer cells. Carcinogenesis. 35(7):1536–1546.
- Rizzo A, Waldner MJ, Stolfi C, Sarra M, Fina D, Becker C, Neurath MF, Macdonald TT, Pallone F, Monteleone G, Fantini MC. 2011. Smad7 expression in T cells prevents colitis-associated cancer. Cancer Res. 71(24):7423–7432.
- Rong X, Ge D, Shen D, Chen X, Wang X, Zhang L, Jia C, Zeng J, He Y, Qiu H, et al. 2018. MiR-27b suppresses endothelial cell proliferation and migration by targeting Smad7 in kawasaki disease. Cell Physiol Biochem. 48(4):1804–1814.
- Sands BE, Feagan BG, Sandborn WJ, Schreiber S, Peyrin-Biroulet L, Frédéric Colombel J, Rossiter G, Usiskin K, Ather S, Zhan X, et al. 2019. Mongersen (GED-0301) for Active Crohnʼs Disease: Results of a Phase 3 Study. Am J Gastroenterol. 115(5):738–745.
- Scarozza P, Schmitt H, Monteleone G, Neurath MF, Atreya R. 2019. Oligonucleotides-A novel promising therapeutic option for IBD. Front Pharmacol. 10:314.
- Sedda S, Franzè E, Bevivino GD, Giovangiulio M, Rizzo A, Colantoni A, Ortenzi A, Grasso E, Giannelli M, Sica GS, et al. 2018. Reciprocal regulation between Smad7 and Sirt1 in the gut. Front Immunol. 9:1854.
- Shen DW, Li YL, Hou YJ, Xu ZD, Li YZ, Chang JY. 2019. MicroRNA-543 promotes cell invasion and impedes apoptosis in pituitary adenoma via activating the Wnt/β-catenin pathway by negative regulation of Smad7. Biosci Biotechnol Biochem. 83(6):1035–1044.
- Shi JQ, Wang B, Cao XQ, Wang YX, Cheng X, Jia CL, Wen T, Luo BJ, Liu ZD. 2020. Circular RNA_LARP4 inhibits the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Eur Rev Med Pharmacol Sci. 24(4):1863–1869.
- Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. 2004. GADD34-PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J Cell Biol. 164(2):291–300.
- Shi X, Chen F, Yu J, Xu Y, Zhang S, Chen YG, Fang X. 2008. Study of interaction between Smad7 and DNA by single-molecule force spectroscopy. Biochem Biophys Res Commun. 377(4):1284–1287.
- Shi Y, Massagué J. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 113(6):685–700.
- Siebel C, Lendahl U. 2017. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 97(4):1235–1294.
- Sim WJ, Iyengar PV, Lama D, Lui SKL, Ng HC, Haviv-Shapira L, Domany E, Kappei D, Tan TZ, Saie A, et al. 2019. c-Met activation leads to the establishment of a TGFβ-receptor regulatory network in bladder cancer progression. Nat Commun [Internet]. [accessed 2020 Sep 9] 10(1):1–19. /pmc/articles/PMC6761206/?report = abstract
- Simonsson M, Heldin CH, Ericsson J, Grönroos E. 2005. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 280(23):21797–21803.
- Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. 2012. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 31(50):5162–5171.
- Song C, Wang J, Ma Y, Yang Z, Dong D, Li H, Yang J, Huang Y, Plath M, Ma Y, et al. 2018. Linc-smad7 promotes myoblast differentiation and muscle regeneration via sponging miR-125b. Epigenetics. 13(6):591–604.
- Song L-Y, Ma Y-T, Wu C-F, Wang C-J, Fang W-J, Liu S-K. 2017. MicroRNA-195 Activates Hepatic Stellate Cells In Vitro by Targeting Smad7. BioMed Res Int. 2017:1–12.
- Song Y, Mou R, Li Y, Yang T. 2020. Zingerone Promotes Osteoblast Differentiation Via MiR-200c-3p/smad7 Regulatory Axis in Human Bone Mesenchymal Stem Cells. Med Sci Monit. 26:e919309
- Souchelnytskyi S, Tamaki K, Engström U, Wernstedt CT, Dijke P, Heldin CH. 1997. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 272(44):28107–28115.
- Stolfi C, De Simone V, Colantoni A, Franzè E, Ribichini E, Fantini MC, Caruso R, Monteleone I, Sica GS, Sileri P, et al. 2014. A functional role for Smad7 in sustaining colon cancer cell growth and survival. Cell Death Dis. 5(2):e1073
- Stolfi C, Marafini I, De Simone V, Pallone F, Monteleone G. 2013. The dual role of Smad7 in the control of cancer growth and metastasis. Int J Mol Sci. 14(12):23774–23790.
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature. 403(6765):41–45.
- Sun H, Peng Z, Tang H, Xie D, Jia Z, Zhong L, Zhao S, Ma Z, Gao Y, Zeng L, et al. 2017. Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-β signaling in and promote progression of hepatocellular carcinoma. Oncogene [Internet]. [accessed 2020 Jun 21] 36(21):2957–2968. /pmc/articles/PMC5444978/?report = abstract
- Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. 2002. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J Biol Chem. 277(42):39919–39925.
- Tabata T, Kokura K, Ten Dijke P, Ishii S. 2009. Ski co-repressor complexes maintain the basal repressed state of the TGF-β target gene, SMAD7, via HDAC3 and PRMT5 . Genes Cells. 14(1):17–28.
- Tang Q, Zou Z, Zou C, Zhang Q, Huang R, Guan X, Li Q, Han Z, Wang D, Wei H, et al. 2015. MicroRNA-93 suppress colorectal cancer development via Wnt/β-catenin pathway downregulating. Tumour Biol. 36(3):1701–1710.
- Tang Y, Liu Z, Zhao L, Clemens TL, Cao X. 2008. Smad7 stabilizes β-catenin binding to E-cadherin complex and promotes cell-cell adhesion. J Biol Chem. 283(35):23956–23963.
- Tang Y, Reissig S, Glasmacher E, Regen T, Wanke F, Nikolaev A, Gerlach K, Popp V, Karram K, Fantini MC, et al. 2019. Alternative splice forms of CYLD mediate ubiquitination of SMAD7 to prevent TGFB signaling and promote colitis. Gastroenterology. 156(3):692–707.e7.
- Tao J, Wang J, Li C, Wang W, Yu H, Liu J, Kong X, Chen Y. 2019. MiR-216a accelerates proliferation and fibrogenesis via targeting PTEN and SMAD7 in human cardiac fibroblasts. Cardiovasc Diagn Ther. 9(6):535–544.
- Tenesa A, Farrington SM, Prendergast JGD, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, et al. 2008. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 40(5):631–637.
- Tijsen AJ, van der Made I, van den Hoogenhof MM, Wijnen WJ, van Deel ED, de Groot NE, Alekseev S, Fluiter K, Schroen B, Goumans M-J, et al. 2014. The microRNA-15 Family Inhibits the TGFβ-pathway in the Heart. Cardiovasc Res. 104(1):61–71.
- Tong L, Chu M, Yan B, Zhao W, Liu S, Wei W, Lou H, Zhang S, Ma S, Xu J, et al. 2017. MTDH promotes glioma invasion through regulating miR-130b-ceRNAs. Oncotarget. 8(11):17738–17749.
- Tong L, Shen S, Huang Q, Fu J, Wang T, Pan L, Zhang P, Chen G, Huang T, Li K, et al. 2020. Proteasome-dependent degradation of Smad7 is critical for lung cancer metastasis. Cell Death Differ. 27(6):1795–1806.
- Topper JN, Cai J, Qiu Y, Anderson KR, Xu YY, Deeds JD, Feeley R, Gimeno CJ, Woolf EA, Tayber O, et al. 1997. Vascular MADs: Two novel MAD-related genes selectively inducible by flow in human vascular endothelium. Proc Natl Acad Sci USA. 94(17):9314–9319.
- Tsai TH, Sun MH, Ho TC, Ma HI, Liu MY, Tsao YP. 2014. Notch prevents transforming growth factor-β-assisted epithelial-mesenchymal transition in cultured limbal progenitor cells through the induction of Smad7. Mol Vis. 20:522–534.
- Tsunobuchi H, Ishisaki A, Imamura T. 2004. Expressions of inhibitory Smads, Smad6 and Smad7, are differentially regulated by TPA in human lung fibroblast cells. Biochem Biophys Res Commun. 316(3):712–719.
- Tu X, Zheng X, Li H, Cao Z, Chang H, Luan S, Zhu J, Chen J, Zang Y, Zhang J. 2015. MicroRNA-30 protects against carbon tetrachloride-induced liver fibrosis by attenuating transforming growth factor β signaling in hepatic stellate cells. Toxicol Sci. 146(1):157–169.
- Tuncer E, Calçada RR, Zingg D, Varum S, Cheng P, Freiberger SN, Deng CX, Kleiter I, Levesque MP, Dummer R, et al. 2019. SMAD signaling promotes melanoma metastasis independently of phenotype switching. J Clin Invest. 129(7):2702–2716.
- Uchida K, Suzuki H, Ohashi T, Nitta K, Yumura W, Nihei H. 2001. Involvement of MAP kinase cascades in Smad7 transcriptional regulation. Biochem Biophys Res Commun. 289(2):376–381.
- Ulloa L, Doody J, Massagué J. 1999. Inhibition of transforming growth factor-β/SMAD signalling by the interferon-gamma/STAT pathway. Nature. 397(6721):710–713.
- Vishal M, Vimalraj S, Ajeetha R, Gokulnath M, Keerthana R, He Z, Partridge NC, Selvamurugan N. 2017. MicroRNA-590-5p stabilizes Runx2 by targeting Smad7 during osteoblast differentiation. J Cell Physiol. 232(2):371–380.
- Von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Böttinger EP. 2000. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor β. J Biol Chem. 275(15):11320–11326.
- Wahl LC, Watt JE, Yim HTT, De Bourcier D, Tolchard J, Soond SM, Blumenschein TMA, Chantry A. 2019. Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms. Int J Mol Sci. 20(19):4682.
- Wang C, Gu S, Cao H, Li Z, Xiang Z, Hu K, Han X. 2016. MiR-877-3p targets Smad7 and is associated with myofibroblast differentiation and bleomycin-induced lung fibrosis. Sci Rep. 6(1):1–11.
- Wang F, Wang J, Yang X, Chen D, Wang L. 2016. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagn Pathol. 11(1):1–9.
- Wang H, Nie L, Wu L, Liu Q, Guo X. 2017. NR2F2 inhibits Smad7 expression and promotes TGF-β-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem Biophys Res Commun. 485(1):181–188.
- Wang J, Zhao J, Chu ESH, Mok MTS, Go MYY, Man K, Heuchel R, Lan HY, Chang Z, Sung JJY, et al. 2013. Inhibitory role of Smad7 in hepatocarcinogenesis in mice and in vitro. J Pathol. 230(4):441–452.
- Wang M, Jia M, Yuan K. 2018. MicroRNA-663b promotes cell proliferation and epithelial mesenchymal transition by directly targeting SMAD7 in nasopharyngeal carcinoma. Exp Ther Med. 16(4):3129–3134.
- Wang M, Saha J, Hada M, Anderson JA, Pluth JM, O'Neill P, Cucinotta FA. 2013. Novel Smad proteins localize to IR-induced double-strand breaks: interplay between TGFβ and ATM pathways. Nucleic Acids Res. 41(2):933–942.
- Wang L, Geng J, Sun B, Sun C, Shi Y, Yu X. 2020. MiR-92b-3p Is Induced by Advanced Glycation End Products and Involved in the Pathogenesis of Diabetic Nephropathy. Evid Based Complement Alternat Med. 2020:6050874.
- Wang R, Fu T, You K, Li S, Zhao N, Yang J, Zhuang SM. 2018. Identification of a TGF-β-miR-195 positive feedback loop in hepatocytes and its deregulation in hepatoma cells. Faseb J. 32(7):3936–3945.
- Wang W, Dong R, Guo Y, He J, Shao C, Yi P, Yu F, Gu D, Zheng J. 2019. CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J Cell Mol Med. 23(8):5486–5496.
- Wang W, Huang XR, Li AG, Liu F, Li JH, Truong LD, Wang XJ, Lan HY. 2005. Signaling mechanism of TGF-β1 in prevention of renal inflammation: Role of Smad7. JASN. 16(5):1371–1383.
- Wang X, Wang H, Cao J, Ye C. 2018. Exosomes from adipose-derived stem cells promotes VEGF-C-dependent lymphangiogenesis by regulating miRNA-132/TGF-β pathway. Cell Physiol Biochem. 49(1):160–171.
- Wang XL, Zhao YY, Sun L, Shi Y, Li ZQ, Zhao XD, Xu CG, Ji HG, Wang M, Xu WR, et al. 2018. Exosomes derived from human umbilical cord mesenchymal stem cells improve myocardial repair via upregulation of Smad7. Int J Mol Med. 41(5):3063–3072.
- Wang Y. 2017. The inhibition of microRNA-15a suppresses hepatitis B virus-associated liver cancer cell growth through the Smad/TGF-β pathway . Oncol Rep. 37(6):3520–3526.
- Wang ZH, Zhang QS, Duan YL, Zhang JL, Li GF, Zheng DL. 2015. TGF-β induced miR-132 enhances the activation of TGF-β signaling through inhibiting SMAD7 expression in glioma cells. Biochem Biophys Res Commun. 463(3):187–192.
- Wei B, Wei W, Zhao B, Guo X, Liu S. 2017. Long non-coding RNA HOTAIR inhibits MIR-17-5p to regulate osteogenic differentiation and proliferation in nontraumatic osteonecrosis of femoral head. PLoS One. 12(2):e0169097.
- Wei Z, Cao J, Zhang X, Yin D, Xu D, Lu G. 2019. EPA Attenuates Epithelial-Mesenchymal Transition and Fibrosis Through the TGF-β1/Smad3/ILK Pathway in Renal Tubular Epithelial HK-2 Cells by Up-Regulating miR-541. Int J Clin Exp Pathol. 12(7):2516–2525.
- Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Ten Dijke P, Chantry A. 2005. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-β signalling. Oncogene. 24(54):8080–8084.
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. 2007. Autoinhibition of the HECT-Type Ubiquitin Ligase Smurf2 through Its C2 Domain. Cell. 130(4):651–662.
- Wojtowicz S, Lee S, Chan E, Ng E, Campbell CI, Di Guglielmo GM. 2020. SMURF2 and SMAD7 induce SARA degradation via the proteasome. Cell Signal. 72:109627
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. 1994. Mechanism of activation of the TGF-β receptor. Nature. 370(6488):341–347.
- Wu Z, Qiu X, Gao B, Lian C, Peng Y, Liang A, Xu C, Gao W, Zhang L, Su P, et al. 2018. Melatonin-mediated miR-526b-3p and miR-590-5p upregulation promotes chondrogenic differentiation of human mesenchymal stem cells. J Pineal Res. 65(1):e12483.
- Xia H, Ooi LLPJ, Hui KM. 2013. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 58(2):629–641.
- Xu HM, Sui FH, Sun MH, Guo GL. 2019. Downregulated microRNA-224 aggravates vulnerable atherosclerotic plaques and vascular remodeling in acute coronary syndrome through activation of the TGF-β/Smad pathway. J Cell Physiol. 234(3):2537–2551.
- Xu J, Ai Q, Cao H, Liu Q. 2015. MiR-185-3p and miR-324-3p predict radiosensitivity of nasopharyngeal carcinoma and modulate cancer cell growth and apoptosis by targeting SMAD7. Med Sci Monit. 21:2828–2836.
- Xu J, Xu Y. 2017. The lncRNA MEG3 downregulation leads to osteoarthritis progression via miR-16/SMAD7 axis. Cell Biosci. 7(1):69.
- Xu P, Lin X, Feng XH. 2016. Posttranslational regulation of smads. Cold Spring Harb Perspect Biol. 8(12):a022087.
- Yan X, Liao H, Cheng M, Shi X, Lin X, Feng XH, Chen YG. 2016. Smad7 protein interacts with receptor-regulated Smads (R-Smads) to inhibit transforming growth factor-β (TGF-β)/Smad signaling. J Biol Chem. 291(1):382–392.
- Yanagida A, Iwaisako K, Hatano E, Taura K, Sato F, Narita M, Nagata H, Asechi H, Uemoto S, Kinoshita M. 2011. Downregulation of the Wnt antagonist Dkk2 links the loss of Sept4 and myofibroblastic transformation of hepatic stellate cells. Biochim Biophys Acta - Mol Basis Dis. 1812(11):1403–1411.
- Yang D, Ma M, Zhou W, Yang B, Xiao C. 2017. Inhibition of miR-32 activity promoted EMT induced by PM2.5 exposure through the modulation of the Smad1-mediated signaling pathways in lung cancer cells. Chemosphere. 184:289–298.
- Yang Y, Ding S, Xu G, Chen F, Ding F. 2017. MicroRNA-15a inhibition protects against hypoxia/reoxygenation-induced apoptosis of cardiomyocytes by targeting mothers against decapentaplegic homolog 7. Mol Med Rep. 15(6):3699–3705.
- Yu D, Shin HS, Lee YS, Lee YC. 2014. MiR-106b modulates cancer stem cell characteristics through TGF-β/Smad signaling in CD44-positive gastric cancer cells. Lab Invest. 94(12):1370–1381.
- Yu F, Guo Y, Chen B, Dong P, Zheng J. 2015. MicroRNA-17-5p activates hepatic stellate cells through targeting of Smad7. Lab Invest. 95(7):781–789.
- Yu J, Lei R, Zhuang X, Li X, Li G, Lev S, Segura MF, Zhang X, Hu G. 2016. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat Commun. 7(1):13884–13812.
- Yu Y, Feng XH. 2019. TGF-β signaling in cell fate control and cancer. Curr Opin Cell Biol. 61:56–63.
- Zeng Z, Yao J, Li Y, Xue Y, Zou Y, Shu Z, Jiao Z. 2018. Anti-apoptosis endothelial cell-secreted microRNA-195-5p promotes pulmonary arterial smooth muscle cell proliferation and migration in pulmonary arterial hypertension. J Cell Biochem. 119(2):2144–2155.
- Zhai W, Li S, Zhang J, Chen Y, Ma J, Kong W, Gong D, Zheng J, Xue W, Xu Y. 2018. Sunitinib-suppressed miR-452-5p facilitates renal cancer cell invasion and metastasis through modulating SMAD4/SMAD7 signals 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. Mol Cancer. 17(1):157
- Zhang B, Zhou M, Li C, Zhou J, Li H, Zhu D, Wang Z, Chen A, Zhao Q. 2014. MicroRNA-92a inhibition attenuates hypoxia/reoxygenation-induced myocardiocyte apoptosis by targeting Smad7. PLoS One. 9(6):e100298.
- Zhang J, Liu W, Shen F, Ma X, Liu X, Tian F, Zeng W, Xi X, Lin Y. 2018. The activation of microRNA-520h–associated TGF-β1/c-Myb/Smad7 axis promotes epithelial ovarian cancer progression. Cell Death Dis. 9(9):1–15.
- Zhang J, Ning X, Cui W, Bi M, Zhang D, Zhang J. 2015. Transforming growth factor (TGF)-β-induced microRNA-216a promotes acute pancreatitis via Akt and TGF-β pathway in mice. Dig Dis Sci. 60(1):127–135.
- Zhang Q. l, Wang W, Li J, Tian SY, Zhang TZ. 2015. Decreased miR-187 induces retinal ganglion cell apoptosis through upregulating SMAD7 in glaucoma. Biomed Pharmacother. 75:19–25.
- Zhang S, Ekman M, Thakur N, Bu S, Davoodpour P, Grimsby S, Tagami S, Heldin CH, Landström M. 2006. TGFβ1-induced activation of ATM and p53 mediates apoptosis in a Smad7-dependent manner . Cell Cycle. 5(23):2787–2795.
- Zhang S, Fei T, Zhang L, Zhang R, Chen F, Ning Y, Han Y, Feng X-H, Meng A, Chen Y-G. 2007. Smad7 antagonizes transforming growth factor β signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 27(12):4488–4499.
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. 2001. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 98(3):974–979.
- Zhang Z, Fan Y, Xie F, Zhou H, Jin K, Shao L, Shi W, Fang P, Yang B, van Dam H, et al. 2017. Breast cancer metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat Commun. 8(1)
- Zhang Z, Jiang H, Li X, Chen X, Huang Y. 2019. MiR‐92a regulates brown adipocytes differentiation, mitochondrial oxidative respiration, and heat generation by targeting SMAD7. J Cell Biochem. jcb.29539.
- Zhang Z, Xue Z, Liu Y, Liu H, Guo X, Li Y, Yang H, Zhang L, Da Y, Yao Z, et al. 2018. MicroRNA-181c promotes Th17 cell differentiation and mediates experimental autoimmune encephalomyelitis. Brain Behav Immun. 70:305–314.
- Zhao F, Ma X, Qiu W, Wang P, Zhang R, Chen Z, Su P, Zhang Y, Li D, Ma J, et al. 2020. Mesenchymal MACF1 Facilitates SMAD7 Nuclear Translocation to Drive Bone Formation. Cells. 9(3):616.
- Zhao S, Sun H, Jiang W, Mi Y, Zhang D, Wen Y, Cheng D, Tang H, Wu S, Yu Y, et al. 2017. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFβ-mediated epithelial to mesenchymal transition. Mol Cancer. 16(1):12
- Zhao T, Sun S, Zhang H, Huang X, Yan M, Dong X, Wen Y, Wang H, Lan HY, Li P. 2016. Therapeutic effects of tangshen formula on diabetic nephropathy in rats. PLoS One. 11(1):e0147693.
- Zhao Y, Thornton AM, Kinney MC, Ma CA, Spinner JJ, Fuss IJ, Shevach EM, Jain A. 2011. The deubiquitinase CYLD targets Smad7 protein to regulate transforming growth factor β (TGF-β) signaling and the development of regulatory T cells. J Biol Chem. 286(47):40520–40530.
- Zhong X, Tang J, Li H, Shi X, Wu Y, Xia D, Zhang H, Ye J, Wu H. 2020. MiR-3175 promotes epithelial-mesenchymal transition by targeting Smad7 in human conjunctiva and pterygium. FEBS Lett. 594(7):1207–1217.
- Zhou G, Lin W, Fang P, Lin X, Zhuge L, Hu Z, Jin L. 2016. MiR-10a improves hepatic fibrosis by regulatingthe TGFβl/smads signal transduction pathway. Exp Ther Med. 12(3):1719–1722.
- Zhou JY, Zheng SR, Liu J, Shi R, Yu HL, Wei M. 2016. MiR-519d facilitates the progression and metastasis of cervical cancer through direct targeting Smad7. Cancer Cell Int. 16(1).
- Zhu B, Wei X, Xia Wang T, Bao Zhou Y, Cai Liu A, Min Zhang G. Wen 2015. Increased miR-16 expression induced by hepatitis C virus infection promotes liver fibrosis through downregulation of hepatocyte growth factor and Smad7. Arch Virol. 160(8):2043–2050.
- Zhu J, Zhang Z, Zhang Y, Li W, Zheng W, Yu J, Wang B, Chen L, Zhuo Q, Chen L, et al. 2018. MicroRNA-212 activates hepatic stellate cells and promotes liver fibrosis via targeting SMAD7. Biochem Biophys Res Commun. 496(1):176–183.
- Zhu M, Zhang N, He S. 2019. Transcription factor KLF4 modulates microRNA-106a that targets Smad7 in gastric cancer. Pathol Res Pract. 215(8):152467.
- Zhu Z, Xu Y, Zhao J, Liu Q, Feng W, Fan J, Wang P. 2015. MiR-367 promotes epithelial-to-mesenchymal transition and invasion of pancreatic ductal adenocarcinoma cells by targeting the Smad7-TGF-β signalling pathway. Br J Cancer. 112(8):1367–1375.
- Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. 2016. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 7(4):e2203–e2203.
- Zorzi F, Calabrese E, Monteleone I, Fantini M, Onali S, Biancone L, Pallone F, Monteleone G. 2012. A phase 1 open-label trial shows that smad7 antisense oligonucleotide (GED0301) does not increase the risk of small bowel strictures in Crohn’s disease. Aliment Pharmacol Ther. 36(9):850–857.
- Zou T, Zhu M, Ma YC, Xiao F, Yu X, Xu L, Ma LQ, Yang J, Dong JZ. 2018. MicroRNA-410-5p exacerbates high-fat diet-induced cardiac remodeling in mice in an endocrine fashion. Sci Rep. 8(1):1–12.
