Abstract
Antimicrobial drug resistance (AMR) from improper use of antibiotics in various livestock products is a growing hazard for humans worldwide, with current death rate in excess of 700,000 per annum linked to the problem. Microorganisms are a rich source of structurally distinct bioactive compounds designed to protect the microbes and can offset AMR challenge. A study was conducted at Chinhoyi University of Technology to isolate, identify and characterize biosurfactant secreting microbes from broiler bird’s gastrointestinal tract. Analysis of variance was performed in Genstat software. 16S rRNA technique was used to identify the DNA of isolates, annotated by similarity using BLASTn analysis against the NCBI nucleotide database. Phylogenetic analysis was performed on the BLASTn outcome to have an appreciation of the evolutionary genetic relationships. Small intestine-derived samples had a wider hemolytic activity of 5.6 mm, with a 39% emulsification index. At 98.29% sequence similarity, the bacterium producing biosurfactants was identified as an Escherichia coli strain similar to the 7.1994/NIST 0056 strain. The biosurfactant substance is a derivative of decane with beta lactams, tetracyclines and sulfa drugs properties which were responsible for the observed antibacterial activity. We recommend endogenous biosurfactant production optimization experiments and in-vivo trials to evaluate the potential impacts of a biosurfactant based feed additive in broilers.
Introduction
Natural products are regarded as important compounds which exhibit many applications in the field of agriculture among others.Citation1 Veterinary medications are widely used in the production of food animals for therapeutic and preventative purposes. If these medications are misused or the advised drug withdrawal times are not followed, residues of these compounds may remain in the animal food products, and the risk they pose to human health cannot be disregarded.Citation2 It is therefore of paramount importance to advocate for products which pose less risk to human health than synthetic products.
Biosurfactants are amphipathic molecules with a wide range of structural variations, biodegradability and less toxicity compared to their synthetic counterparts.Citation1 Several biosurfactants capabilities include antibacterial, antifungal, and antiviral properties, making them useful molecules in the fight against a variety of diseases and infections.Citation3–5 Biosurfactants can also be used as anti-adhesive agents against infections. In this way, the release of biosurfactants by probiotic bacteria in-vivo is a defence weapon against other colonization strains in the gastrointestinal tracts.Citation5,Citation6 Microbes can survive a wide range of stressful situations thanks to their diverse biosurfactant properties, allowing them to conquer a wide range of settings. These functions can be exploited and used to alter the productivity of broiler chickens.
The demand for developing clean, nontoxic, and environmentally friendly synthetic approaches (green chemistry) in the synthesis of various agricultural compounds that will end up in the environment has increased in response to the world’s growing environmental concerns.Citation7 The creation of advanced bio-based materials is the result of this. Synthetic surfactants can be replaced with microbial surfactants (biosurfactants), which also have potential applications in the biomedical, industrial, and environmental fields as well as being potential antibiotic agents.Citation1 Low toxicity, improved environmental compatibility, high selectivity, and specific activity at extremely adverse conditions like pH, temperature, and salinity are just a few of the intriguing characteristics of biosurfactants.Citation7 These factors make biosurfactants potential candidates for a variety of uses, such as their use in the field of animal sciences. Depending on the microorganisms that produce them, various types of biosurfactants exhibit a wide range of physiological functions and have different properties.Citation8 Hydrophobic compound solubilization, heavy metal binding, virulence factors, cell signaling (quorum sensing), and biofilm formation are noteworthy among all of these characteristics. However, an accurate characterization of the compounds and any potential toxic side effects is necessary for their potential application in a variety of products, such as feed additives. This study presents isolation, identification and characterization of endogenous biosurfactants secreted by a gut extracted microbe in broilers fed a commercial diet with the objective of counteracting antimicrobial resistance challenge.
Materials and methods
Study site
The study was conducted at Zimbabwe’s Chinhoyi University of Technology (CUT) biotechnology laboratory located 17.3533° S and 30.2058° E. CUT farm bred broiler chickens utilizing a deep litter system with wheat straw as bedding were used. The farm is situated at an altitude of 1140 meters in a sub-humid tropical setting. The average annual rainfall at the farm site is 850 mm, and the average daily temperature ranges from 7 °C in winter to 27 °C in summer. Cambisols which are granite-derived soils are present at the farm.Citation9
Sample collection
The birds, from which samples were taken, were taken care of in accordance with the established regulations in ‘The governance of animal care and use for scientific purposes in Africa and the Middle East’.Citation10 Birds were kept under similar conditions to those under which commercial farm animals are kept hence authorization from an Ethics Committee were not necessary as per directive no. 2010/63/EU of the European Parliament and of the Council.
The gastrointestinal tract (GIT) of healthy chickens which were fed a commercial diet was collected at slaughter age of six weeks and transported on ice to the laboratory. The GIT was sectioned into four tissues: crop, gizzard, small and large intestines. The digesta samples from each tissue section were then aseptically collected into 2 mL Eppendorf tubes for immediate culturing. Collected GIT samples were serially diluted (up to 10−5) in 0.85% ringer solution. Serial dilutions of each region were spread plated in triplicate on nonselective multi-nutrient agar medium (mass/volume: 0.5% peptone, 0.3% beef extract, 1.5% agar) and incubated at 30 °C for 24 h. The plate count method, as published by the,Citation11 was used to determine the microbial population. After the incubation phase, colony forming units (cfu) were determined using a colony counter.
Isolation of potential biosurfactant producing bacteria
The hemolytic activity, oil spread, and oil drop collapse assays were used to identify potential biosurfactant secreting bacteria.
Hemolytic activity
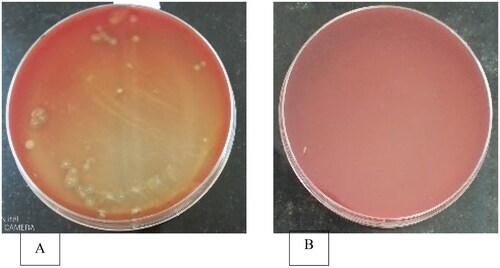
The method described by Walter et al.Citation12 was followed. Blood agar base was prepared according to the manufacturer’s directions, then sterile sheep blood at a rate of 5% v/v was added at a temperature of 55 °C in a water bath. The liquid mixture was gently stirred until the blood was evenly dispersed, and then it was poured aseptically into petri plates. Sheep blood was used in the blood agar because of its’ increased sensitivity to the hemolytic toxins released by bacterial cells thus causing hemolytic zones around the colonies over the period of time.Citation13
Bacterial colonies which were grown on nonselective multi-nutrient agar medium were transferred to 5% (v/v) sheep blood agar using a sterile Whatman filter paper size 1 (in place of velveteen membrane). First, the Whatman filter paper was cut into circular disks which can fit inside a petri dish with a flip of paper on one end to facilitate easy lifting of the paper. The cut Whatman filter papers were then steam autoclaved for sterilization purposes. Colony lifting was done aseptically in a lamina flow. A velveteen membrane was carefully placed on top of nutrient agar plates that had been incubated at 30 °C for 24 h. The membrane copied microbial cells which were then transferred onto blood agar plates, where they were gently pushed to imprint the cells onto the blood agar. For 48 h, the cultures were incubated at 30 °C. The appearance of a clear zone surrounding the bacterial colonies indicated hemolytic action.Citation13 After testing for hemolytic activity, colonies with clear zones on blood agar were streaked and subcultured on multi-nutrient agar. This was done to ensure that the cultures were pure.
Following the hemolysis test, pure colonies were transferred to a multi-nutrient broth medium and cultured for six days at 30 °C. Biosurfactants were extracted from supernatant by centrifugation at 5 000 rpm for 20 min. The supernatant was collected for additional screening tests, that is the oil spread method, oil drop collapse, and emulsification activity.Citation14
Oil spreading method
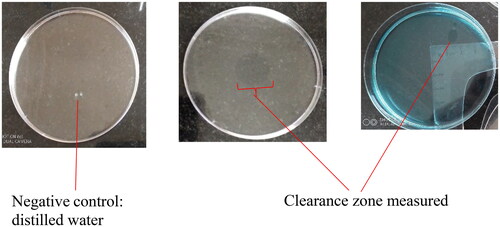
Distilled water (40 mL) was poured on to the petri dishes, followed by addition of vegetable oil (10 µL) placed to the center of the petri dish. Following that, a drop of cell-free culture broth supernatant (10 µL) was put over the vegetable oil surface. The diameter of the clearance zone on the oil surface was measured and compared to that of the negative control (10 µL of distilled water).Citation14
Oil drop collapse method
The method described by Jain et al.Citation15 was followed. In each petri plate, ten microlitres of vegetable oil were put. After that, 10 µL of cell-free culture broth were added, and the drop on the oil surface image was examined after 2 m. When the cultures produced a flat drop, this was deemed positive biosurfactant production. Isolates that produced round droplets were scored as negative, indicating a lack of biosurfactant production.Citation12
Complementary screening of biosurfactant producing bacteria
Microbial isolates that were positive for at least one primary screening method were subjected to an emulsification capacity assay as a supplement to confirm their potential to secrete biosurfactants. An emulsification index (E24) devised by Cooper and GoldenbergCitation16 was used to assess the emulsifying potential of isolated strains. Six milliliters of vegetable oil were mixed with four milliliters of culture supernatant. For 2 m, the liquid was vortexed at high speed to fully combine the supernatant and oil. The combination was left to stand for 24 h. The E24 index was derived by dividing the height of the emulsified layer (mm) by the overall height of the liquid column (mm) (i.e., height of oil + emulsion layer).Citation12,Citation17 The results were compared to distilled water, which served as a negative control.Citation14
Identification of biosurfactant secreting bacterial isolates
A method described by Rayeni and NezhadCitation14 was followed. Ten milliliters of an overnight culture of selected microbe, in multi-nutrient broth, was added to 500 mL of multi-nutrient broth and incubated for 7 days at 30 °C. To recover the biosurfactant, the bacteria were eliminated by centrifuging at 5000 rpm for 20 m. The pH of the supernatant was adjusted to 2 with 6 N HCI and then the solution was stored at 4 °C for 24 h to precipitate the biosurfactants. The biosurfactant within the organic layer were obtained by vigorously mixing a solution of chloroform and methanol (2:1 v/v) with the precipitated biosurfactants. Solvents were opted for due to their extensive capacity to solubilize a number of secondary metabolites. Also, methanol contains both polar and nonpolar groups which make it able to extract both polar and nonpolar compounds. Mixing ensures movement of biosurfactants from the hydrophilic phase (nutrient broth) into the organic, hydrophobic phase.Citation14 This layer was separated using a separating funnel and dried at 50 °C for 4–5 h to obtain dry mass which was then taken for further analysis using a GC/MS and FTIR spectroscopy to determine chemical components. The biosurfactant samples were first screened for antibiotic properties using radio receptor assay at the Residue Analysis Division of the Central Veterinary Laboratory (CVL), Harare. The assay detects substances in the sample that have characteristics similar to a particular antibiotic drug. Biosurfactants were then evaluated for their in-vitro antibacterial properties against Escherichia coli and Staphylococcus aureus using the agar well diffusion method.Citation18 Studies were conducted using distilled water as a negative control and Terranox (positive control), which contained oxytetracycline soluble powder at a recommended rate of 2 mg per 1 mL of water. The zones of growth inhibition (mm) around the disks were measured after 24 h of incubation at 37 °C. Obtained data were analyzed using a one-way analysis of variance (ANOVA) in Genstat 18th edition and means were separated using Fischer’s least significant difference at 5% confidence interval.
Isolates positive for the biosurfactant production screening tests were taken for identification using the 16S rRNA technique. Bacterial DNA was extracted from the culture using the Quick-DNA™ Fungal/Bacterial Minirep Kit (ZYMO RESEARCH, Catalogue No. D6005) according to the manufacturer’s protocol. The concentration and purity of the extracted DNA was determined using an absorption spectroscopy model at wavelengths 260 nm and 280 nm. The integrity of the PCR amplicons was visualized on 1% agarose gel (CSL-AG500, Cleaver Scientific Ltd) stained with EZ-vision® Bluelight DNA Dye. The NEB Fast Ladder was used on all gels (N3238) as size standard. The 16S gene target region (27–1492 bp) was amplified using the universal primer sets from Inqaba Biotechnology, South Africa: 16S-27F5′ – AGAGTTTGATCMTGGCTCAG −3′ and 16S-1492R 5′ – CGGTTACCTTGTTACGACTT −3′.Citation19 The PCR was realized on a thermal cycler under the following conditions: NEB OneTaq 2x MasterMix with standard buffer (Catalogue No. M0482S), Genomic DNA (10–30 ng/µl), Forward primer (10 µM), Reverse primer (10 µM), and Nuclease free water (Catalogue No. E476). Amplification was performed using the initial denaturation at 94 °C for 5 m, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, extension at 68 °C for 1 m, and a final extension at 68 °C for 10 m.
The amplicons were enzymatically purified using the ExoSAP procedure (NEB M0293L; NEB M0371), for sequencing (zymo Research, ZR-96 DNA Sequencing Clean-up Kit™, Catalogue No. D4050), and sequenced in the forward and reverse direction (Nimagen, BrilliantDye™ Terminator cycle sequencing Kit V3.1, BRD3-100/1000) using the ABI 3730x/Genetic analyzer (Applied Biosystems, Thermo Fisher Scientific).
FinchTV (https://finchtv.software.informer.com/1.4/) was used to view the raw chromatogram files (.abi). CLC Bio Main Workbench was used to assemble the forward and reverse sequencing reads to form a consensus sequence for each sample. BLASTn analysis (with default parameters) was performed against NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine if a sequence in the database matches the query sequence above a certain threshold (99% query coverage; 99% identity).
Similarity scores from BLASTn analysis were extracted into a binary table (https://doi.org/10.6084/m9.figshare.23574249.v1) for phylogenetic diversity analysis between the identified strains of bacterial species. Phylogenetic analysis was done using the Unweighted Pair Group Method (UPGM) of cluster analysis in MultiVariate Statistical Package (MVSP) version 3.22 Kovach Computing Services.
Results
Broiler birds GIT microbial distribution map
There were variations in microbial populations along the gastrointestinal tract of birds, with significant differences between the proximal gut compartments (crop and gizzard) and distal compartments (intestines) ().
Table 1. Average colony forming units per milliliter for each GIT section.
The crop and gizzard had the same microbial populations which were different from those from the small and large intestines.
Hemolytic activity
Hemolytic activity was detected as the presence of clear zones around the bacterial colonies (). More microorganisms with greater clearance zones were found in the intestines, with small intestine derived samples having a wider clearance. The smallest clearance zone was seen in the gizzard and crop.
More colony forming units from the large intestines, as well as the small intestines, demonstrated hemolytic activity. However, small intestine derived samples had a larger clearance zone (5.6 mm) in the current study, followed by large intestines which was significantly different (p < 0.05) from those of the crop and gizzard ().
Table 2. Distribution of microbes showing clearance zones in sheep blood agar.
Colonies with larger clearance zones were subcultured to obtain pure cultures. These colonies had a variety of characteristics, with some exhibiting a slower development rate as indicated by a smaller colonized area ().
Table 3. Phenotypic characteristics of bacterial colonies subcultured from broiler birds’ GIT extracted samples.
Oil spreading method
Distilled water did not show any clearance activity unlike the biosurfactant containing supernatant (). The presence of biosurfactants in the supernatant will result in repulsion of the oil on water surface. This happens as a result of the amphiphilic properties of biosurfactants, thus a clearance activity confirms the availability of the biosurfactants in the supernatant.
With substantial clearance from small intestine samples, the gizzard did not demonstrate clearance zone ().
Table 4. Oil spread test clearance zones and average emulsification index (E24) for biosurfactants from different sections of broiler GIT.
Clearance zones for oil spread technique were significantly different among all the tissues sampled. Larger clearing zones in intestine-derived samples may point to increased competition for resources among the occupants, as well as potentially powerful biosurfactants from this region.
Emulsification capacity (E24) of broiler GIT extracted biosurfactants
The extractions showed some emulsification capabilities proving useful as biosurfactants. On the E24 index, crop and large intestine derived samples trailed small intestine derived samples ().
Annotation by similarity using BLASTn
The DNA material was pure and of one type as indicated by a single strand on the gelCitation20 thus the sample was good for sequencing. Similarity between the sequence queried and the biological sequences within the NCBI database was performed and a 98.29% identity was reviewed pointing to the E. coli strain 7.1994 as the microbe similar to the one producing the biosurfactants in the current study (https://doi.org/10.6084/m9.figshare.23574249.v1).
Genetic diversity of the biosurfactant secreting bacterial strains
A dendrogram () was produced from phylogenetic analysis done using the Unweighted Pair Group Method (UPGM) of cluster analysis in MultiVariate Statistical Package (MVSP) version 3.22 Kovach Computing Services.
Figure 3. Dendogram of community relatedness of forty-nine bacterial strains based on the Jaccard’s coefficient.
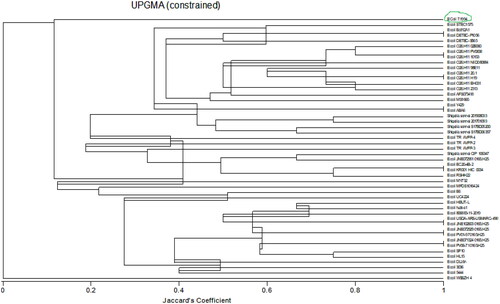
A total of forty-nine bacterial strains, consisting of 44 E. coli and 5 Shigella sonnei strains, which had a higher similarity score from BLASTn results were analyzed.
Gas chromatography mass spectrometry (GCMS)
The endogenous biosurfactants from E. coli strain 7.1994 related microbe showed the presence of Decane (C10H22), octadecane (C18H38), furane-2-carboxylate (C11H7FO3), and 1,7-Di-4-nitroheptan (C21H23NO6) phytochemical compounds with matching scores above 90% ().
Table 5. Major phytochemical compounds identified in biosurfactant GCMS analysis.
Fourier-transform infrared (FTIR) spectroscopy
The extracted biosurfactants have peaks in the triple (2000–2500 cm−1), double (1500–2000 cm−1) and fingerprint regions (600–1500 cm−1) ().
Figure 4. A comparison of FTIR spectroscopy of Pseudomonas aeruginosa (bottom) and crude endogenous biosurfactant (top) extracted from E. coli strain. NB. Pseudomonas aeruginosa adapted from Khademolhosseini et al.Citation21
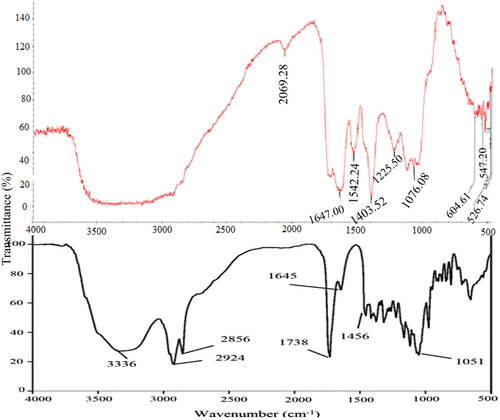
Possible causes of observed peaks are presented in . A molecule’s ability to undergo the numerous reactions required to sustain life is provided by the carbonyl compound it contains, which occurred at 2069.28 wavenumber.
Table 6. FTIR peaks identified in Experimental biosurfactant.
Screening biosurfactants for presence of antibiotics properties
Tetracyclines and sulfa drug properties in the extracted biosurfactant sample were found to be present ().
Table 7. Small intestine extracted biosurfactant sample radio receptor assay output.
Betalactams, which represent penicillin, produced negative results together with amphenicols. Tetracyclines and sulfa drugs were positive probably because of their continued use in broilers.
Antibacterial activity of broiler birds’ GIT extracted biosurfactants
The clearance zones were more noticeable in samples from the small intestine, with the positive control having the highest clearance ().
Table 8. Observed broiler GIT derived biosurfactants antimicrobial activity at 5% (m/v).
Similar to the positive control, the biosurfactants from the small intestine had distinct inhibition zones. For biosurfactants extracted from the small intestine, minimum inhibition activity was seen at a concentration of 5%.
Discussion
The GIT microbial distribution map shows that even when birds are fed the same meal and exposed to the same environmental conditions, there were differences in the amount of microorganisms in their GIT. Choi et al.Citation22 observed similar findings that were attributed to changes in time between feeding and sampling. In birds used in the current study, the large intestines contained more microbes than the small intestines. However, the intestines had less microbial population than the crop and gizzard, (p < 0.05). Each GIT section has distinct metabolic responsibilities that influence the microbial profile,Citation23 hence varying microbial population densities were observed. However, same range populations in the crop and gizzard, as supported by Fathima et al.Citation6 (p. 4), could be attributed to a shorter time period between feeding of the birds and sampling time. However, the intestines had less microbial populations which could be attributed to the acidic conditions of the proventriculus which can reduce populations of less acid adaptable microbes as the digesta passes through. More microorganisms in the large intestines may be credited to a stable environment because the digesta would have traveled through the preliminary GIT portions and some nutrients would have been absorbed. Also, because the quality of the digesta reaching the large intestines is partially uniform and rich in fibrous substance in the diet,Citation24 the bacteria are not subjected to successive changes in the chemical composition of the digesta. The recent findings support Bailey’sCitation25 arguments that the abundance and variety of the microbiota vary along the GIT, with fewer numbers of bacteria in locations with less acceptable circumstances and faster passage of gut contents. The gizzard has muscular walls and grit that serve primarily to physically crush the ingested feed. There were significant differences between birds in the crop microbial community. Some birds will pick on litter material regardless of whether they are fed ad libitum or not. This litter material may include a diverse microbial community, and because the crop will be the first port of call, variances in microbial communities amongst birds in the same area can exist.
Microbial variations observed in the current study in various parts of the same bird gut cements the findings of Oakley and Kogut,Citation26 who proposed that location in the GIT, among other things, influences the composition of microbial populations. As diverse as they appear in the various portions of the GIT, it follows that a vast range of bacterial metabolites with various roles are released into the system of birds. If the crop and the gizzard’s narrower hemolytic clearing zones are any indication, they are related to their primary responsibilities, which do not necessitate a diverse array of activities. The crop moistens the feed in preparation for the physical activity of the gizzard. However, in the small intestines, there is a higher microbial activity,Citation25 and other organisms such as E. coli live there. A greater clearance zone suggested that bacteria from this region were producing more biosurfactants.
The current clearance zone on oil spread is less than what was reported by Alkan et al.Citation17 when they worked with lactic acid bacteria strains which ranged from 1.87 to 5.92 cm. Eighty percent of nutrient absorption occurs in the upper section of the small intestine,Citation27 therefore, biosurfactants from this region are potential biofilm disruptors and beneficial metabolites because they are most likely produced by nutrient utilization related microorganisms. The oil drop collapse method did not produce results within the timed interval of 2 min. However, cultures gave a flat drop after the cut off time. This was partly attributed to a low concentration of biosurfactants in the extracts used, hence in the current study, this was not a reliable screening technique.
Some microorganisms have hemolysis characteristics but do not release biosurfactants outside of their cells.Citation28 This will result in more microorganisms being reported on hemolysis activities, however on emulsification tests, the biosurfactants’ emulsification ability will be severely reduced because the test will be done by the biosurfactants rather than the microbes. Nayarisseri et al.,Citation29 avers that samples with more than 30% emulsification activity indicates a greater activity. The emulsification values obtained in the current study were in the same range reported by Alkan et al.Citation17 when they worked with lactic acid bacterial species which showed emulsification capabilities in the range 19.5–58%. The tests performed in the current study show that the small intestine derived sample has higher activity. The findings augment assertions by Sambanthamoorthy et al.,Citation30 that biosurfactants have important qualities, among them substantial emulsification activity. Small intestine samples showed an upper hand and were examined further for their antibacterial qualities to see if they can suppress the growth and development of common infections under stressful settings.
The phylogenetic dendrogram shows the genetic evolutionary relationships or similarity proportion between the identified bacterial strains.Citation31 The dendrogram shows two major groups, E. coli strain WSSZH4 and the other strains emanating from the other group. This points to the fact that E. coli strain WSSZH4 is distinct from all the other forty-eight strains studied in this dendrogram. This is supported by Devanga Ragupathi et al.Citation32 who affirms that there are some distinct strains of E. coli which are not related to other strains of E. coli or Shigella strains. Shigella is a genus of Gram-negative bacteria genetically closely related to E. coli.Citation33 Some of its characteristics include it being a facultative anaerobic, non-spore-forming, nonmotile, and rod-shaped bacterium.Citation34 The dendrogram shows that the Shigella strains are closely related to some strains of E. coli, particularly E. coli strain STEC 1575 and E. coli strain JNE072951 0165:H25. Since the E. coli and Shigella can occur together in the gut of broilers and have a common score in this study, there is a chance that these microbes exchanged their genetic material between the involved strains, equipping the E. coli strain with the biosurfactant production genes. Shad and ShadCitation35 have reported the ability of cross-immunisation by some other strains of Shigella in the gut. This partly explains the biosurfactant production ability of the E. coli strain in the current study. The dendrogram affirms the BLASTn analysis output as indicated by an E. coli strain 7.1994 (green encircled on dendrogram) with a Jaccard’s coefficient less than 0.2, meaning less than 20% similarity with the rest of the other strains picked in the BLASTn analysis. The current findings point to an E. coli bacterium closely related to E. coli strain 7.1994/NIST 0056 bacterium being responsible for biosurfactant production in the gut of broilers.
A GCMS match factor score above 90 is considered an excellent match, 90 is a good match, 70–80 is a fair match, and 60% is a poor match.Citation36 As a result, the biosurfactant compounds discovered in the current study fall into the excellent category, with Octadecane receiving a highest score of 97.4% match factor. Octadecane, a hydrophobic molecule, is a straight chain alkane carrying 18 carbon atoms. It has a role as a bacterial metabolite and a plant metabolite.Citation37 Decane (CAS# 124-18-5) is an aliphatic hydrocarbon primarily derived from crude oil. In addition to the rubber and paper industries, it is used as a solvent in organic syntheses. Furane-2-carboxylate (4′-Fluorophenyl) has been reported as a factor with potential inhibitory activity against bacterial swarmingFootnote1 and likely inhibit extracellular polysaccharide production.Citation38 The inhibitory activity seen in the current study may be due to trace amounts of this compound, which would give the secreting microbe immunity. This is a positive development for the creation of a novel anti-infective strategy to reduce the overuse of synthetic drugs in broiler production, as supported by Rütschlin and Böttcher.Citation39 1,7-Di(2′-methoxyphenyl)-4-nitroheptan-1,7-dione may be associated with control of inflammation, wound and muscular atrophy, and immune disorders as put forward by Shih et al..Citation40 This chemical may be useful in microbe defence mechanisms against certain drugs.
Other biosurfactants have been reported to have peaks in the regions noted in the current study.Citation41,Citation42 However, no peaks were observed above the 2100 wavenumbers (cm−1). Regarding the number of peaks, there are seven peaks, informing that the analyzed biosurfactant is a small organic component as supported by Nandiyanto et al.Citation43 There is evidence of functional groups in the extracted biosurfactants as evidenced by absorbance bands above 1500 cm−1 (). Since there were no peaks in the 3650–3200 wavenumber range, the sample had dried completely and had not absorbed any chemotherapeutic water.Citation43 A carbonyl compound gives a biological molecule the ability to generate new molecules and be altered with a variety of other functional groups. Because they are polar in nature, carbonyl compounds have minute positive and negative charges.
The hydrophilic and hydrophobic regions of chemically produced surfactants are clearly separated into a hydrophilic head group (charged or polar) and a hydrophobic tail, which is made up of linear alkyl groups.Citation44 The observed peak indicates the presence of amides or carboxylates functional groups in the double bond region because it occurred below 1700 cm−1.Citation43 Peaks in the range of 1670–1620 cm−1, as highlighted by Nandiyanto et al.,Citation43 represent unsaturation bonds (double and triple bond). As a result, an unsaturated bond accounts for the observed peak at 1647 wavenumbers. It is possible that the benzene ring, which produced a peak at wavenumber 1542, is attributable to this alkene stretch. This nitrogen-oxy compound supports the discovery of the aliphatic hydrocarbon decane (CAS 124-18-5) made by the GCMS. The biosurfactant possesses polar properties and some antibiotic-like ionizable compounds (at 1076.08 wavenumbers).Citation45
Tetracyclines and sulfonamides, picked by the radio receptor assay, are two antibiotic classes of popular veterinary use for animals.Citation2,Citation45 Therefore, it stands to reason that some intestinal microbes may have evolved specific mechanisms to produce similar compounds for their defence. However, beta lactams (penicillins) are the preferred medication for humans,Citation46 so their use in animals is constrained. This partially explains the low concentrations of these substances because microbes lack the ability to adjust to and become accustomed to the synthesis of such substances. Biosurfactants have tremendous therapeutic potential and antimicrobial qualities, and they can accomplish their intended functions with fewer side effects. The current study results support the hypothesis put forth by Sambanthamoorthy et al.Citation30 that biosurfactants had stronger antibacterial properties. Growth inhibition zones were observed around the biosurfactant extracts, but not around the negative control (distilled water). On samples from the crop and large intestines, antibacterial activity was, however, barely detectable. Staphylococcus aureus responded more to biosurfactants addition. The results of the study support the claims made by Naughton et al.Citation47 that biosurfactants can have a number of functions, including antibacterial activity. The negative control (distilled water) had the greatest number of colony forming units as it failed to suppress microbial growth. Escherichia coli exhibited some resistance to the crop’s biosurfactants. However, there was significant clearance around the biosurfactants retrieved from the intestines.
Conclusion and recommendations
A biosurfactant-secreting microbe was isolated from broiler GIT and identified as E. coli strain 7.1994. The potential use of the biosurfactants from this strain of E. coli in antimicrobial resistance challenge lies in its exhibited oil spreading activity and a higher emulsification index, 39%, in vegetable oil. The FTIR and GCMS spectra show that the biosurfactant is a glycolipid, derivative of decane without hydrate component but with double and triple bonds related to ketones. Therefore, this E. coli strain 7.1994 and its biosurfactant could be suitable for use in the fight against antimicrobial resistance challenge from broiler products. Researchers recommend further studies on optimization of biosurfactant release and in-vivo trials with broiler chickens.
Ethics approval
The birds were taken care of in accordance with the established regulations in “The governance of animal care and use for scientific purposes in Africa and the Middle East”. Birds were kept under similar conditions to those under which commercial farm animals are kept hence authorization from an Ethics Committee were not necessary as per directive no. 2010/63/EU of the European Parliament and of the Council.
Acknowledgements
The researchers are grateful for the support offered by Chinhoyi University of Technology Biotechnology laboratory technicians, Great Zimbabwe University Biology Laboratory Technicians, Inqaba biotech South Africa for genome analyses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data are presented in-text, and a link has been provided for data in repository.
Additional information
Funding
Notes
1 Swarming is a coordinated form of motility in which bacteria can migrate in groups of flagella-driven cells over semi-solid surfaces (46).
References
- Garg M, Priyanka, Chatterjee M. (2018). Isolation, characterization and antibacterial effect of biosurfactant from Candida parapsilosis. Biotechnol Rep, 18, e00251.
- Planche C, Chevolleau S, Noguer-Meireles MH, et al. Fate of sulfonamides and tetracyclines in meat during pan cooking: focus on the thermodegradation of sulfamethoxazole. Molecules. 2022;27(19):6233.
- Elisa MdO, Victor HGS, Marcelo SA, Jerri ÉZ, Wardsson LB, Tiago MdS Isolation and characterization of biosurfactant-producing bacteria from Amapaense Amazon soils. Int J Microbiol. 2021;2021:1–11.
- Gayathiri E, Prakash P, Karmegam N, Varjani S, Awasthi MK, Ravindran B. Biosurfactants: potential and eco-friendly material for sustainable agriculture and environmental safety—a review. Agronomy. 2022;12(3):662.
- Rocha V, Teixeira JA, Rodrigues LR. Antimicrobial and antiadhesive properties of a biosurfactant isolated from Lactobacillus paracasei ssp. paracasei A20. Lett Appl Microbiol. 2010;50(4):419–424.
- Fathima S, Shanmugasundaram R, Adams D, Selvaraj RK. Gastrointestinal microbiota and their manipulation for improved growth and performance in chickens. Foods. 2022;11(10):1401.
- Kumar CG, Mamidyala SK, Das B, Sridhar B, Devi GS, Karuna MSL. Synthesis of biosurfactant-based silver nanoparticles with purified rhamnolipids isolated from Pseudomonas aeruginosa BS-161R. J Microbiol Biotechnol. 2010;20(7):1061–1068.
- Díaz De Rienzo MA, Stevenson P, Marchant R, Banat IM. Antibacterial properties of biosurfactants against selected Gram-positive and -negative bacteria. FEMS Microbiol Lett. 2016;363(2):fnv224.
- Mashavakure N, Mashingaidze AB, Musundire R, et al. Influence of tillage, fertiliser regime and weeding frequency on germinable weed seed bank in a subhumid environment in Zimbabwe. S Afr J Plant Soil. 2019;36(5):319–327.
- Mohr BJ, Fakoya FA, Hau J, Souilem O, Anestidou L. The governance of animal care and use for scientific purposes in Africa and the Middle East. ILAR J. 2016;57(3):333–346.
- Association of Official Analytical Chemists. 1990. Animal feed. In: Helrich K, ed. Encyclopedia of Grain Science. 15th ed., Vol. 1, Issue Volume 1. Gaithersburg, MD: Association of Official Analytical Chemists.
- Walter V, Syldatk C, Hausmann R. Screening concepts for the isolation of biosurfactant producing microorganisms. Adv Exp Med Biol. 2010;672:1–13.
- Microbeonline. Blood agar: Composition, preparation, uses and types of hemolysis - microbeonline. https://microbeonline.com/blood-agar-composition-preparation-uses-and-types-of-hemolysis/. Published 2017.
- Rayeni LT, Nezhad SS. Characterization of biosurfactant produced by probiotic bacteria isolated from human breast milk. Int J Basic Sci Med. 2018;3(1):18–24.
- Jain DK, Collins-Thompson DL, Lee H, Trevors JT. A drop-collapsing test for screening surfactant-producing microorganisms. J Microbiol Methods. 1991;13(4):271–279.
- Cooper DG, Goldenberg BG. Surface-active agents from two Bacilllus species. Appl Environ Microbiol. 1987;53(2):224–229.
- Alkan Z, Ergİnkaya Z, Konuray G, Turhan EÜ. Production of biosurfactant by lactic acid bacteria using whey as growth medium. Turk J Vet Anim Sci. 2019;43(5):676–683.
- Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6(2):71–79.
- Mahlangu SG, Tai SL. Morphological and molecular characterization of bacterial endophytes from Centella asiatica leaves. J Genet Eng Biotechnol. 2022;20(1):171.
- Wittmeier P, Hummel S. Agarose gel electrophoresis to assess PCR product yield: comparison with spectrophotometry, fluorometry and qPCR. Biotechniques. 2022;72(4):155–158.
- Khademolhosseini R, Jafari A, Mousavi SM, Hajfarajollah H, Noghabi KA, Manteghian M. Physicochemical characterization and optimization of glycolipid biosurfactant production by a native strain of Pseudomonas aeruginosa HAK01 and its performance evaluation for the MEOR process. RSC Adv. 2019;9(14):7932–7947.
- Choi JH, Kim GB, Cha CJ. Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult Sci. 2014;93(8):1942–1950.
- Yeoman CJ, Chia N, Jeraldo P, Sipos M, Goldenfeld ND, White BA. The microbiome of the chicken gastrointestinal tract. Anim Health Res Rev. 2012;13(1):89–99.
- Bailey RA. Gut health in poultry - the world within. https://www.thepoultrysite.com/articles/gut-health-in-poultry-the-world-within. Published 2013.
- Bailey RA. Gut health in poultry: the world within - update. Published 2019. https://www.thepoultrysite.com/articles/gut-health-in-poultry-the-world-within-1.
- Oakley BB, Kogut MH. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front Vet Sci. 2016;3:11.
- Ravindran V, Abdollahi M. Nutrition and digestive physiology of the broiler chick: state of the art and outlook. Animals. 2021;11(10):2795.
- Patowary K, Patowary R, Kalita MC, Deka S. Characterization of biosurfactant produced during degradation of hydrocarbons using crude oil as sole source of carbon. Front Microbiol. 2017;8(279):279.
- Nayarisseri A, Singh P, Singh SK. Screening, isolation and characterization of bio-surfactant producing Bacillus subtilis strain ANSKLAB03. Bioinformation. 2018;14(6):304–314.
- Sambanthamoorthy K, Feng X, Patel R, Patel S, Paranavitana C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BioMed Central Microbiol. 2014;14:1–9.
- Miller JT, Jolley-Rogers G, Mishler BD, Thornhill AH. Phylogenetic diversity is a better measure of biodiversity than taxon counting. J Syst Evol. 2018;56(6):663–667.
- Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Inbanathan FY, Veeraraghavan B. Accurate differentiation of Escherichia coli and Shigella serogroups: challenges and strategies. New Microbes New Infect. 2018;21:58–62.
- Kinavouidi DJ, Kayath CA, Nieko NPMD, Mokemiabeka SN, Moukala DC, Nguimbi E. Shigella strain has developed non-studied pathogenicity mechanisms of adaptability in the colonization of epithelial cells. AiM. 2022;12(04):270–294.
- Shi R, Yang X, Chen L, et al. Pathogenicity of Shigella in chickens. PLoS One. 2014;9(6):e100264.
- Shad AA, Shad WA. Shigella sonnei: virulence and antibiotic resistance. Arch Microbiol. 2021;203(1):45–58.
- Jordi Labs. 2017. NIST/EPA/NIH Mass spectral library compound scoring: match factor, reverse match factor, and probability. Jordi Labs, 1–2. https://jordilabs.com/wp-content/uploads/2017/07/Whitepaper-NIST-EPA-NIH-Mass-Spectral-Library-Compound-Scoring.pdf.
- National Center for Biotechnology Information. PubChem compound summary for CID 11635, octadecane. https://pubchem.ncbi.nlm.nih.gov/compound/Octadecane. Published 2022.
- Hara S, Isoda R, Tahvanainen T, Hashidoko Y. Trace amounts of furan-2-carboxylic acids determine the quality of solid agar plates for bacterial culture. PLoS One. 2012;7(7):e41142.
- Rütschlin S, Böttcher T. Inhibitors of bacterial swarming behavior. Chemistry. 2020;26(5):964–979.
- Shih C, Shi Q, Wang H. Compounds with (1E, 6E)-1,7-bis-(3,4-dimethoxyphenyl)-4-4-distributed-hepta-1,6-diene-3,5-dione structural scaffold, their biological activity and uses thereof: Vol. US 9,562,0. https://patentimages.storage.googleapis.com/58/32/98/7b304d7d1ac0c5/US9562025.pdf. Published 2017.
- Essghaier B, Mallat N, Khwaldia K, Mottola F, Rocco L, Hannachi H. Production and characterization of new biosurfactants/bioemulsifiers from Pantoea alhagi and their antioxidant, antimicrobial and anti-biofilm potentiality evaluations. Molecules. 2023;28(4):1912.
- Kiran GS, Priyadharsini S, Sajayan A, Priyadharsini GB, Poulose N, Selvin J. Production of lipopeptide biosurfactant by a marine Nesterenkonia sp. and its application in food industry. Front Microbiol. 2017;8:1138.
- Nandiyanto ABD, Oktiani R, Ragadhita R. How to read and interpret FTIR spectroscope of organic material. Indonesian J Sci Technol. 2019;4(1):97–118.
- Otzen DE. Review. Biosurfactants and surfactants interacting with membranes and proteins: same but different? Biochim Biophys Acta Biomembr. 2017;1859(4):639–649.
- Hu J, Tang X, Qi M, Cheng J. New models for estimating the sorption of sulfonamide and tetracycline antibiotics in soils. Int J Environ Res Public Health. 2022;19(24):16771.
- Egorov AM, Ulyashova MM, Rubtsova MY. Inhibitors of β-lactamases. New life of β-lactam antibiotics. Biochemistry. 2020;85(11):1292–1309.
- Naughton PJ, Marchant R, Naughton V, Banat IM. Microbial biosurfactants: current trends and applications in agricultural and biomedical industries. J Appl Microbiol. 2019;127(1):12–28.