Abstract
Sperm mRNA transcriptional profiling can be used to evaluate the fertility of breeding bulls. The aim of the study was to compare the modified RNA isolation methods for higher RNA yield and quality from freshly ejaculated sperm of cattle and buffalo bulls. Ten fresh ejaculates from each Sahiwal (n = 10 bulls × 10 ejaculates) and Murrah bulls (n = 10 bulls x 10 ejaculates) were used for RNA isolation. From the recovered live sperm, total sperm RNA was isolated by conventional methods (TRIzol, Double TRIzol), membrane-based methods combined with TRIzol (RNeasy + TRIzol) with the addition of β-mercaptoethanol (BME) and Kit (RNeasy mini) methods in fresh semen. Among different isolation methods; the membrane-based modified methods combined with TRIzol (RNeasy + TRIzol) with the addition of β-mercaptoethanol (BME) resulted significantly (p < .05) higher total RNA quantity (300–340 ng/µL) and better purity in different concentrations of spermatozoa viz., 30–40 million, 70–80 million and 300–400 million sperm. The study concluded that the inclusion of BME to the combined membrane-based methods with somatic cell lysis buffer solution was best for constant increased yield and purity of RNA isolation from Sahiwal cattle and Murrah buffalo bull sperm.
Introduction
Semen evaluation is an important criteria for successful cryopreservation and artificial insemination. Traditionally, several methods were developed to evaluate semen quality in the laboratory, like assessing genomic DNA integrity, acrosome, plasma membrane, mitochondria, and sperm-oocyte interactions. However, creating a consistent technique for routine RNA isolation from Sahiwal cattle and Murrah buffalo bull sperms to develop a noninvasive methodology to evaluate bull fertilizing ability is limited. Sperm RNA transcripts are important in spermatogenesis, sperm maturation, fertilization, oocyte genome activation, embryogenesis, and placental development, as well as the events surrounding capacitation, motility, and fertilization.Citation1–3 The development of new methods such as mRNA profiling of bull sperm and understanding the significance of these transcripts would be helpful to study the fertilizing capacity of the sperm. Several investigations in humans sperm reported that spermatozoa contains more than 3000 mRNAs that reflect the gene expression during spermatogenesis.Citation4–9
Earlier it was believed that spermatozoal RNAs are transcriptionally inactive; however, latest investigations reported that spermatozoa transport specific functional RNAs into the oocyte at the time of fertilization and these RNAs contribute to early embryonic development.Citation10–16 The process transcription is thought to be terminated after mid-spermiogenesis, and mature spermatozoa do not exhibit considerable transcriptional activity. Interestingly, distinct messenger RNA populations have been discovered in bull spermCitation17 and the existence of spermatozoa RNA has been established in mammals,Citation10,Citation17–19 poultryCitation20 and boar.Citation21,Citation22 The levels of expression of specific sperm transcripts have been linked to sperm functional characteristics,Citation11,Citation23,Citation24 early embryonic development,Citation25 and fertility.Citation26,Citation27 The spermatozoa transcripts represent the spermatogenic process and have predictive value in fertilization, they may be employed as a helpful marker for high-fertile bulls.Citation10,Citation11,Citation28 To obtain precise sperm transcript expression, it is necessary to isolate sufficient quantities and quality RNA from sperm. In sperm, there is less cytoplasm, fewer full-length RNAs with physiologically degraded RNAs, and severely condensed DNA due to protamination.Citation29 Because of the complexity of RNA, spermatozoa require a different RNA isolation technique than other cells. To isolate total RNA, the sperm membrane and nucleo-protamine complex should be totally dissolved. Various protocols to isolate sperm RNA with varying results have been used in different species i.e., human,Citation4,Citation11,Citation30,Citation31 bovine,Citation10,Citation17,Citation27,Citation32,Citation33 porcineCitation26,Citation34 stallionCitation28 and chicken.Citation20 The lack of a suitable species-specific sperm RNA separation procedure has hindered research on sperm RNAs in the majority of species and appropriate and accurate method for sperm RNA isolation and quality assessment in Sahiwal and Murrah buffalo bull is not yet optimized. The isolation sperm RNA meets numerous challenges, specifically low RNA quantity (22–45 times less RNA than haploid spermatid) and the requirement of somatic cells removal.Citation5,Citation17,Citation30 Furthermore, sperm are extremely compacted cells with variances in sperm characteristics and chromatin packaging among species, making cellular content isolation difficult.Citation35 Besides, RNAs exist in the nucleus in a highly fragmented state in spermatozoa.Citation36
Although useful procedures for spermatozoa input were reported, an appropriate and accurate method for RNA measurement and quality assessment was not specified in detail.Citation10,Citation32,Citation33 As there are few conventional protocols for sperm RNA isolation procedures from Sahiwal cattle and Murrah buffalo bulls, this study will be key for developing a sperms RNA isolation procedure, obtaining high-quality RNA for subsequent molecular research, and presenting an effective conclusion.Citation37 The purpose of this study was to develop a suitable protocol for obtaining high-quantity andquality RNA in indigenous cattle and buffalo in a species-specific manner for transcriptome analysis, regardless of seasonal variation or ejaculate quality.
Material and methods
The chemicals used in the experiments were procured from Sigma-Aldrich, St. Louis, MO, USA, Thermo Fisher Scientific, Waltham, Massachusetts, United States and Qiagen, Hilden, Germany.
Experimental animal
The healthy, sexually matured Sahiwal (n = 10) and Murrah buffalo (n = 10) bull (4–5 years of age) maintained at the Artificial Breeding Research Center (ABRC), ICAR-National Dairy Research Institute (Deemed University) in Karnal, Haryana, India which were in regular semen collection were selected for the study.
Study location
The experiment was executed at the Artificial Breeding Research Center, ICAR-National Dairy Research Institute (Deemed University) in Karnal, Haryana, India. The Institute is located at 29.43°N latitude and 72.2 °C longitudes, at 250 meters above mean sea level. In the summer, the highest ambient temperature reaches 45 °C, while in the winter, it drops to around 2 °C. During July and August, the area receives 760–960 mm of rain, with relative humidity ranging from 41 to 85 percent.
Semen collection and storage
Preputial washing of the bulls was carried out with normal saline one day before the semen collection. The bulls were thoroughly washed, cleaned, and dried at least 20 min before semen collection in the early morning. The semen sample was taken twice a week using an artificial vagina (IMV; L'Aigle, Cedex, France) set at 41 °C with a 15–20 min gap between two consecutive collections.
A total of 100 fresh ejaculates from each Sahiwal (n = 10 × 10) and Murrah bulls (n = 10 × 10) irrespective of seasons throughout the year. The bulls were fed with concentrate ration and seasonal green fodder as per ICAR (2013) recommendations. For quality ejaculates, timely vaccination, de-worming, a regular checkup for other infectious diseases, and a routine herd-health program was followed. The fresh ejaculates were evaluated for ejaculate volume, mass motility, and initial progressive motility. Ten ejaculates were collected from each bull based on initial motility (>70%) and mass activity 3+ or higher, and one mL of neat semen was collected from each ejaculate in a 1.5-mL microcentrifuge tube for further processing.
Sample preparation for RNA isolation
The RNA isolation area was prepared to be clean and decontaminated using RNaseZAP® spray. The following criteria were used for total RNA extraction and purification procedureCitation38 Free of protein (absorbance 260 nm/280 nm); undegraded (28S:18S ratio) should be roughly between 1.8 and 2.0 and accepted as pure RNA, Free of genomic DNA. The expected 260 nm/230 nm ratio are generally in the range of 2.0–2.2, Free of nucleases for extended storage, RNase free glass and plasticware, The RNase and DNase free chemicals and plasticware were used for RNA isolations.
Sperm recovery and separation
Swim-up technique
Swim-up technique for live sperm separation and recovery was followed as per the method defined by previous researcher.Citation39 In brief 1 mL of freshly ejaculated semen sample was layered gently below the pre-loaded 1 mL of equilibrated sp-TALP medium in a 15 mL centrifuge tube to recover the motile spermatozoa. After preparation of the different layers (Bottom-semen sample, and Top-sp-TALP), the tubes were incubated at 39 °C. After the incubation for the period of 1 h, 750 µL of the upper fraction of the sp-TALP layer was aspirated, resuspended with 3 mL Sp-TALP medium, and centrifuged for 10 min at 300 × g. Finally, the sperm pellets were resuspended in 100 µL Sp-TALP, and the sperm recovery was estimated using a photometer (IMV, L'Aigle, France). The recovered motile sperm were varied from 300 to 400 × 106 sperm/mL and the same concentration of sperm was used for the total RNA isolation.
Somatic cell removal
To ensure the purity of the sperm RNA, removal of the somatic cells must be considered during the RNA isolation procedures. A Somatic cell lysis buffer (SCLB) containing 0.1% SDS, 0.5% Triton (X-100) was used to lyse somatic cells for sperm cell purification.Citation3,Citation4,Citation30,Citation40,Citation41
Sperm concentration for total RNA isolation
The sperm concentrations of 30–40 million, 70–80 million and 300–400 million sperm cells were used for RNA isolation and comparison of different isolation methods to select the best method for achieving required quantity with quality of total sperm RNA from bull spermatozoa.
Isolation of total sperm RNA
The total sperm RNA was isolated by different methods viz., conventional methods TRIzol,Citation42 Double TRIzol, membrane-based methods combined with TRIzol (TRIzol + RNeasy) with the addition of β-mercaptoethanol (BME)Citation43 and pure kit methods with slight modifications.
Conventional methods
TRIzol method
The pre-prepared sperm pellet was mixed with 1 mL of PBS, and washed three times by centrifugation at 3000 rpm for 5 min at 4 °C for purification of the sperm cell from other somatic cells, germ cells and leukocytes. According to the manufacturer’s instructions, total sperm RNA was extracted from bull sperm using TRIzol (Invitrogen, USA) with minor modifications.
In a short, 0.5 mL of ice-cold TRI reagent was added to the sperm pellet and homogenized 25–30 times with a 20-G needle attached to a 5-mL syringe. After vortexing for 5 min, the samples were incubated at room temperature (RT) for 5 min until the sperm membrane was completely dissociated. 200 µl chloroform was added to the lysate and thoroughly shaken by hand for at least 20 s, followed by 5 min at room temperature. The mixture was centrifuged at 12,000 × g for 20 min at 4 °C to separate the phases. After centrifugation, three layers were observed viz. the upper aqueous layer (RNA), middle white layer (DNA) and bottom pink layer (protein). The RNA-containing upper aqueous layer was transferred to a new 1.5 mL conical microcentrifuge tube, where an equal volume of isopropanol was added and gently mixed by reversing the tubes. The mixture was kept for 10 min at RT and centrifuged at 12,000 g for 15 min at 4 °C. Supernatant was discarded after centrifugation and 1 mL of 99.99% ethanol was added to the RNA pellet and again centrifuged at 12,000 g for 10 min. Finally, ethanol was removed, and the RNA pellet was air-dried. The pellet was dissolved in 40 µl of nuclease-free water, followed by the addition of 2 µl of RNase inhibitor (20 U/l, Invitrogen) and 10 mM dithiothreitol (DTT).
Double TRIzol method
The prepared pellet was resuspended in 1 mL of PBS, and washed three times by centrifugation at 3000 rpm for 5 min at 4 °C to purify the spermatozoa by removing the other somatic cells, germ cells and leukocytes.
TRIzol reagent was used to extract total RNA from spermatozoa. In this procedure the samples were lysed twice with TRIzol reagent for complete dissociation of sperm membrane. In short, 0.5 mL of ice-cold TRIzol was added to sperm pellet and homogenized for a minimum of 25–30 times with a 20-G needle attached to a 5-mL syringe. The samples were then vortexed for 5 min at room temperature and incubated for 5 minutes, centrifuged at 12,000 g for 30 s, 0.5 mL of fresh, chilled TRIzol was added to the supernatant and vortexed for 1–2 min, and the rest of the procedure was carried out as described in the TRIzol method.
Combined method
RNeasy + TRIzol method with β-mercaptoethanol (BME)
The pre-prepared pellet was mixed with 1 mL of PBS, and the sperm sample was washed three times by centrifugation at 3000 rpm for 5 min at 4 °C to remove the somatic and other germ cells.
Here, the total sperm RNA was isolated by combining the conventional and kit (RNeasy) methods described by earlier workerCitation30 with modifications (). Briefly, the sperm pellet was homogenized with a 20-G needle for about 25–30 times in 1 mL SCLB containing - β mercaptoethanol (10 µl/mL). The homogenized mixture was vortexed for 1–2 min and incubated at RT for 30 min. 0.5 mL of ice-chilled TRI reagent was added to the mixture and again subjected to vortexing for 2 minutes. The mixed sample was then maintained at room temperature for 15 min without being disturbed before adding 200 µl cooled chloroform. The contents were mixed vigorously by hand for 20 s and kept at RT for 3–5 min, followed by centrifugation at 12,000 g for 20 min at 4 °C. The upper aqueous layer was aspirated into a sterilized 2.0-mL tube, and an equal volume of 100% ethanol was pipetted in and mixed thoroughly. The mixture was transferred to a mini-spin column provided and processed as per the manufacturer’s protocol. Three times elution was done at room temperature with different volumes (30, 25 and 15 µl) of nuclease-free water. Finally, the three elutes were pooled, and 10 mM DTT was added to the pooled RNA, which was stored at −80 °C until further processing.
RNeasy + TRIzol method without β-mercaptoethanol
The pre-prepared sperm pellet was resuspended in 1 mL of PBS, and the sperm sample in PBS was centrifuged three times for 5 min at 3000 rpm at 4 °C to separate the spermatozoa from somatic cells, germ cells, and leukocytes, as described above.
Total sperm RNA was isolated using a modified combination of conventional and RNeasy methods. BME was not used in this method, unlike the previous one.Citation30
Membrane-based method
RNeasy mini kit method
The pre-prepared sperm pellet was resuspended in 1 mL of PBS and washed three times by centrifugation at 3000 rpm for 5 min at 4 °C to remove other somatic cells, germ cells, and leukocytes from the spermatozoa.
The QIAGEN RNeasy® Mini Kit (Cat No./ID: 74104) was used to isolate the total RNA as per the manufacturer’s instructions. To eliminate genomic DNA, RNAse-free, DNAse I treatment (Qiagen) was performed.
Estimation of total RNA yield and quality
RNA quality estimation
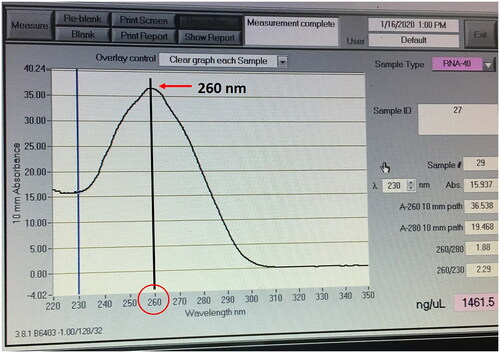
Estimated total RNA concentration and the optical density ratios (OD- 260/280 and OD- 260/230) with nono drop spectrophotometer (ND-1000, Thermo Scientific, USA). The absorbance at 230, 260 and 280 nm was used to determine the purity of isolated RNA. The 260/280 ratio was used to determine potential protein contamination, whereas the 260/230 ratio was used to determine salt and organic solvent contamination. The absorbance at 260 nm was measured with 1 µL of RNA sample, and the total RNA concentration was represented in nanograms per microliter (ng/µl) units. A bioanalyzer was used to examine the RNA fragment size distribution and peak (Agilent 2000, Agilent Technologies, USA).
Gel electrophoresis
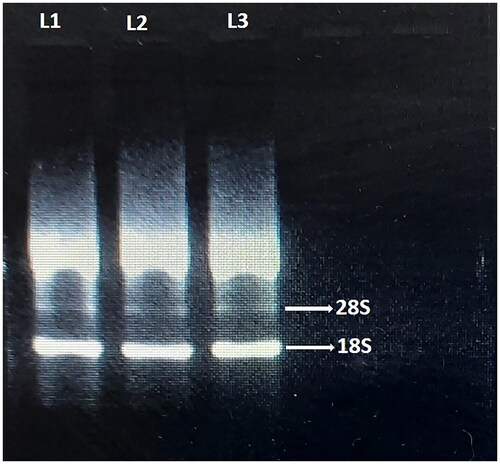
The quality of the isolated RNA was observed by gel electrophoresis using 2.0% agarose gel stained with ethidium bromide to visualize the double band rRNA. Electrophoretic methods have been applied to separate the samples according to the size of comprised molecules to evaluate the degree of degradation. Reliable RNA integrity is evaluated using agarose gel electrophoresis stained with ethidium bromide, which produces a specific band pattern.Citation44 Gel image shows double bands containing the 28S and 18S ribosomal RNA (rRNA) and other bands. When the ratio of 28S:18S bands is within 1.8–2.0 at 260/280 nm, the RNA sample is considered as high quality.
Statistical analysis
The data recorded were analyzed using the SPSS software (version 22). One-way analysis of variance (ANOVA) followed by Tukey’s post hoc test to compare the means and determine the significant differences between the groups were used to select the input spermatozoa concentration and the protocol used for RNA isolation based on total RNA yield and quality. The values were presented as means ± SE, and p < .05 was considered to be statistically significant.
Results
Sperm purification and recovery
A swim-up technique was applied to the freshly ejaculated semen sample to recover the motile spermatozoa. The recovered motile sperm was 300–400 × 106 sperm/mL and different concentrations of sperm viz., 30–40, 70–80 and 300–400 million sperm cells were used for the RNA isolation. The results of total RNA yield and the purity of isolated RNA using different protocols are described below-
Total sperm RNA yield
According to spectrophotometric measurement, total RNA yield in Sahiwal and Murrah buffalo bull spermatozoa was considerably (p < .05) higher in the TRIzol + RNeasy procedure with the addition of BME in 300–400 million sperm concentrations than in all other methods under consideration ( and ).
Table 1. Total RNA yield of Sahiwal bull spermatozoa using different isolation methods (N = 100) (Mean ± SE).
Table 2. Total RNA yield of Murrah bull spermatozoa using different isolation methods (N = 100) (Mean ± SE).
The TRIzol + RNeasy + BME method resulted in the highest total RNA yield of 39.24 ± 0.41 ng/µL, 68.87 ± 1.75 ng/µL and 340.72 ± 19.31, ng/µL in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively as compared to other isolation methods in Sahiwal bull sperm. It is followed by the TRIzol + RNeasy method, double TRIzol method, TRIzol, and RNeasy (). Similar results were found in the Murrah bull sperm with the highest total RNA yield in TRIzol + RNeasy + BME method resulting 51.12 ± 2.16 ng/µL, 72.23 ± 2.65 ng/µL and 318.20 ± 37.60 ng/µL in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively followed by TRIzol + RNeasy method, double TRIzol method, TRIzol and RNeasy ().
Sperm RNA quality
The TRIzol + RNeasy with the addition of the BME method yielded good quality RNA without contaminating substances than the other methods in both Sahiwal and Murrah bull spermatozoa ( and ), the peak at 260 mm absorbance (). In Sahiwal bull sperm, the isolated RNA's purity was best in the TRIzol + BME + RNeasy method with 1.95 ± 0.00, 1.93 ± 0.01 and 1.90 ± 0.01 in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively followed by TRIzol + RNeasy, RNeasy, TRIzol and Double TRIzol (). Similar trends also resulted in Murrah bull sperm. The best quality RNA was found in TRIzol + BME + RNeasy method with 1.97 ± 0.00, 1.91 ± 0.01 and 1.98 ± 0.00 in 30–40 × 106, 70–80 × 106 and 300–400 × 106, respectively followed by TRIzol + RNeasy, RNeasy, TRIzol and Double TRIzol ().
Table 3. RNA purity of Sahiwal bull spermatozoa using different isolation methods (N = 100) (Mean ± SE).
Table 4. RNA purity of Murrah bull spermatozoa using different isolation methods (N = 100) (Mean ± SE).
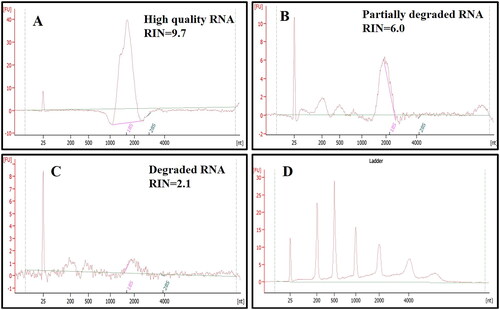
The Agilent 2100 Bio-Analyzer was used for RNA integrity number (RIN) to estimate the purity or integrity of RNA. Based on the peaks, the RNA integrity was measured by RNA integrity number value. RIN values are measured from 1 to 10, where RIN Value 1–5 indicates complete degradation and 5–7 shows partially degraded RNA and RIN value above 7 indicates high quality RNA. In our study, the RIN value of 2.1–9.7 was observed in different samples, indicating the RIN of 9.7, highly intact and 2.1 as highly degraded (). Persistence of 18S and/or 28S rRNA showing the isolated RNA integrity (). The double band rRNA was visualized by running 2.0% agarose gel electrophoresis ().
Discussion
In the present study, we considered the motile and spermatozoa free from other contaminating and somatic cells. The total RNA yield was highest in the modified method (TRIzol + RNeasy + BME), i.e., 39.24 ± 0.41 ng/µL, 68.87 ± 1.75 ng/µL and 340.72 ± 19.31, ng/µL in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively as compared to other isolation methods in Sahiwal bull sperm. It was followed by the TRIzol + RNeasy method, double TRIzol method, TRIzol, and RNeasy (). Similar trends were found in the Murrah bull sperm with the highest total RNA yield in TRIzol + RNeasy + BME method resulting 51.12 ± 2.16 ng/µL, 72.23 ± 2.65 ng/µL and 318.20 ± 37.60 ng/µL in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively followed by TRIzol + RNeasy method, double TRIzol method, TRIzol and RNeasy. The same method displayed the best optical density (OD) at A260/A280 ratio of 1.97, 1.93 and 1.90 in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively in Sahiwal bull sperm, and 1.97, 1.91 and 1.98 in 30–40 × 106, 70–80 × 106 and 300–400 × 106 sperm input concentration, respectively in Murrah bull sperm, which indicates that samples free from protein and organic-free substances among the different methods used. In earlier studies,Citation3 mammalian spermatozoa gene expression profiling has been projected as a novel noninvasive tool to evaluate male fertility. Regardless of the acute contribution of sperm RNA toward the male fertility,Citation5,Citation45 embryo development,Citation30 epigenetic inheritance for acquired traits involved in the paternal genomeCitation46 and health,Citation47 the scarcity of optimization of transcriptomic investigation outfits prevents a thorough understanding of sperm biology. This research aimed to recommend a methodology for extracting high-quality total RNA from bull spermatozoa for transcriptome analysis. High-quality total RNA from bull spermatozoa can be used for transcriptomic analysis. RNA sequencing data can be used to identify markers for bull fertility. It is suggested that analysis of RNAs derived from spermatozoa could provide possible links between the sperm proteome and sperm freezing.Citation12,Citation21,Citation48–51 Because of reduced cytoplasm, a low number of intact and physiologically degraded RNAs,Citation11 a high proportion of gDNA (genomic DNA), and protamination of the nucleus in sperm, optimizing an RNA separation process for bovine sperm is crucial.Citation30 Studies demonstrated that cryopreservation protocol, alter transcripts considered as spermatozoa quality markers and markers for pregnancy success. Most of the studied transcripts considered as male quality markers (PRM1, PRM2, and PEG1/MEST) and reported that mRNAs as markers of pregnancy success (ADD1) were reduced after cryopreservation.Citation52 Because the accuracy of functional genomics research is determined by RNA quality and inter-species variability in sperm characteristics and packaging, sperm RNA separation procedures must be adapted to each species ( and ).
Table 5. RIN values of Sahiwal bull spermatozoa using different isolation methods (N = 100) (Mean ± SE).
Table 6. RIN values of Murrah bull spermatozoa using different isolation methods (N = 100) (Mean ± SE).
Even after the purification of spermatozoa by suitable sperm separation techniques, a major challenge is to extract RNA of sufficient quantity and high quality RNA (i.e., non-degraded RNA and decontaminated). Sperm needs to be purified using various methods for making sperm RNA free from other cell contaminants, such as swim-up protocol,Citation34,Citation53 percoll gradient solutionCitation17,Citation33 and somatic cell lysis.Citation3,Citation11,Citation31 The different procedures of sperm separation and semen storage condition might influence the sperm recovery and the total RNA yield.Citation29,Citation54
Throughout the sperm epididymal maturation, sperm protamines undergo thiol oxidation intra-molecular form followed by inter cell disulfide bonds.Citation55 The covalent sulfur-sulfur (S-S) bonds stabilize the sperm DNA and are believed critical to condense the mammalian sperm nucleus into its fully mature form. During the period of RNA isolation, utmost care should be adopted to avoid degradation by RNases enzymes. Intracellular RNases are released during the RNA isolation procedure, particularly at the lysis step and must be rapidly and thoroughly inactivated to obtain high-quality RNA. β-mercaptoethanol is deliberated a reducing agent that conclusively denatures RNases by disrupting the disulfide bonds and by removing the innate conformation required for enzyme action. Any RNases present in the sample to be extracted will be completely deactivated when combined with the strong denaturing properties of guanidiumisothiocyanate (GITC) and RLT buffer supplied in the RNeasy kits.Citation56 The addition of β-mercaptoethanol can act as a biological antioxidant by scavenging hydroxyl radicals and might be resulted in a high yield of RNA because of its property to break the disulfide bonds (S-S) and additional lysis and nuclear component dissociation. Since membranes provided in the kit can hold only RNA, repeated washing procedures eliminate other contamination like proteins, salts, and inorganic solvents.
As a major source of bacteria found in soil, bedding, and manure, the bull’s preputial orifice is one of the most important sourcesCitation57 and to minimize the bacterial load from the ejaculates before collection of the semen; preputial washing was done with normal saline. Preputial wash helps yield good quality RNA since the ejaculates collected for RNA extraction were free from any contamination like somatic cells, etc. Somatic cell lysis buffer is one of the potent buffer which helps in the lysis of any somatic cells. The composition of somatic cell lysis buffer and the time the sperm cells are exposed to lysis buffer are also very important while isolating spermatozoal RNA, since more prolonged exposure removes the somatic cells, yielding high quality RNA profiles in sperm samples. RNA quantity and quality were found to be far better in the case of TRIzol associated with membrane-based methods (TRIzol + RNeasy), supported by earlier reportsCitation43,Citation58 when compared to membrane-based RNA isolation methods alone. The TRIzol reagent protects RNA integrity while breaking cells and dissolving cell components during the homogenization or lysis process. The quality of RNA was better in the membrane-based kit method resulted in our study also in agreement with previous reports.Citation20,Citation59 Even though the overall structure of spermatozoa is similar across species, very combative procedures must be required to disrupt the membrane structure of bovine spermatozoa. By heating the lysis buffer supplied in the Qiagen kit, human sperm RNA samples were able to extract.Citation3,Citation4 This method was ineffective for bull spermatozoa because extensive cellular debris is deposited at the bottom of the tube after the lysis process, resulting in low total RNA quantity. Indeed, mature spermatozoa are not known to be transnationally active, implying that rRNAs required for ribosome assembly may not be present.
Five techniques have been attempted and compared to recommend a suitable method for bovine sperm RNA isolation. When membrane-based approaches combined with TRIzol (TRIzol + BME + RNeasy) were compared to other methods, both the amount and quality of recovered RNA were higher (TRIzol, Double TRIzol and RNeasy). Our study’s findings agree with earlier researchersCitation43 who isolated RNA from Holstein Friesian bulls and reported higher RNA yield in RNeasy + TRIzol method compared with the other methods. Higher concentration (37.39 ± 1.04 ng/µL) was found in the present study from 30 to 40 million sperm as compared to the previous research (15.22 ± 0.27 ng/µL in 30 million sperm) in Murrah Buffalo bull sperm using heated TRIzol + RNeasy mini method with PVP-coated silica colloidal solution (PVA-Si; Percoll).Citation60 Similarly another study reported using TRIzol + RNeasy method with an average RNA yield of 8.15 ng/µL from 70–80 million sperm in Gir bull, which was found to be lower than the present investigation 68.87 ± 1.75 ng/µL and 72.23 ± 2.65 in Sahiwal and Murrah bull sperm, respectively.Citation61 This might be due to β-mercaptoethanol effectively dissolved the nucleoprotamine complex and unfolded the nuclear proteins, resulting in improved nuclear component lysis and dissociation and the elimination of gDNA in the combined lysis procedures. Additionally, the purity results found in the kit method, which is membrane-based, specifically holds the only RNA; hence, the RNA's purity was superior with no/less gDNA in the membrane-based techniques. Moreover, the repeated washing procedure removed the salts and organic solvents in the membrane-based methods and demonstrated a peak at 260 nm using a spectrophotometer. The RIN value and the peaks observed in the present study with the value of 2.1–9.7 was pragmatic in different samples and are also following the earlier reports.Citation61,Citation62
Conclusions
The findings suggest that the membrane-based methods with TRIzol and somatic cell lysis buffer, BME or DTT, and for generating good quality and quantity RNA from fresh bovine and buffalo bull spermatozoa, further phase separation is necessary and highly effective. We have observed consistent RNA yield and quality from the 100 ejaculates from both cattle and buffalo bull in our findings irrespective of seasonal variation of semen quality which may help further transcriptome analysis and other molecular studies of semen biology. This is the report of consistent of RNA yield and quality from fresh ejaculated semen from Sahiwal cattle and Murrah bull irrespective of seasonal variation of semen quality. However, further optimization of RNA yield and quality from frozen semen is required from indigenous cattle and buffalo.
Authors contributions
RKD: Methodology, Writing- Original draft preparation, Formal analysis. TKM: Conceptualization, Resources, Funding acquisition, Methodology, Supervision, Investigation, Reviewing and Editing, Final approval of the version to be submitted. SN: Methodology, Investigation, Formal analysis, Reviewing and Editing. MB: Investigation, Supervision, Reviewing and Editing. HPY: Sample collection, Reviewing and Editing. RKB: Supervision.
Ethical approval
The animal study was reviewed and approved by The Institutional Animal Ethics Committee (IAEC, ICAR- National Dairy Research Institute, Karnal, Haryana, India) approved all experimental procedures with approval ID-43-IAEC-18-21. Permission to collect semen sample was obtained from the authorized person in Research Center for bull by verbal. All procedures were handled strictly in accordance with the good animal practice to minimize animal sufferings during semen sampling process.
Acknowledgments
The authors sincerely acknowledge the Director cum Vice-Chancellor, ICAR-National Dairy Research Institute, Karnal, Haryana, India, to provide the necessary facilities to carry out this work. The authors are thankful to Dr. A. K. Mohanty, Principal Scientist and Dr. T. K. Datta, Principal Scientist, Animal Biotechnology Centre, ICAR-National Dairy Research Institute for providing NanoQuant and NanoDrop spectrophotometer for quantification of the RNA yield and thankful to Dr. S. De, Principal Scientist and Head, Animal Biotechnology Centre for his valuable suggestion and guidance for the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets supporting the conclusions of this study are available from the corresponding author.
Additional information
Funding
References
- Selvaraju S, Parthipan S, Somashekar L, et al. Current status of sperm functional genomics and its diagnostic potential of fertility in bovine (Bos taurus). Syst Biol Reprod Med. 2018;64(6):484–501.
- Sarıozkan S, Bucak MN, Tuncer PB, Büyükleblebici S, Canturk F. Influence of various antioxidants added to TCM-199 on post-thaw bovine sperm parameters, DNA integrity and fertilizing ability. Cryobiology. 2014;68(1):129–133.
- Ozturk AE, Bodu M, Bucak MN, et al. The synergistic effect of trehalose and low concentrations of cryoprotectants can improve post-thaw ram sperm parameters. Cryobiology. 2020;95:157–163.
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360(9335):772–777.
- Ostermeier GC, Goodrich RJ, Diamond MP, Dix DJ, Krawetz SA. Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil Steril. 2005a;83(6):1687–1694.
- Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J Androl. 2005b;26(1):70–74.
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005;6(8):633–642.
- Carreau S, Lambard S, Said L, Saad A, Galeraud-Denis I. RNA dynamics of fertile and infertile spermatozoa. Biochem Soc Trans. 2007;35(Pt 3):634–636.
- Galeraud‐Denis I, Lambard S, Carreau S. Relationship between chromatin organization, mRNAs profile and human male gamete quality. Asian J Androl. 2007;9(5):587–592.
- Cappallo-Obermann H, Schulze W, Jastrow H, Baukloh V, Spiess AN. Highly purified spermatozoal RNA obtained by a novel method indicates an unusual 28S/18S rRNA ratio and suggests impaired ribosome assembly. Mol Hum Reprod. 2011;17(11):669–678.
- Hamatani T. Human spermatozoal RNAs. Fertil Steril. 2012;97(2):275–281.
- Card CJ, Anderson EJ, Zamberlan S, Krieger KE, Kaproth M, Sartini BL. Cryopreserved bovine spermatozoal transcript profile as revealed by high-throughput ribonucleic acid sequencing. Biol Reprod. 2013;88(2):49–41.
- Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA. Reproductive Medicine Network. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update. 2013;19(6):604–624.
- Bucak MN, Keskin N, Bodu M, et al. Combination of trehalose and low boron in presence of decreased glycerol improves post-thawed ram sperm parameters: A model study in boron research. Andrology. 2022;10(3):585–594.
- Bucak MN, Akalın PP, Keskin N, et al. Combination of fetuin and trehalose in presence of low glycerol has beneficial effects on freeze-thawed ram sperm. Andrology. 2021;9(3):1000–1009.
- Bucak MN, Keskin N, İli P, et al. Decreasing glycerol content by co-supplementation of trehalose and taxifolin hydrate in ram semen extender: microscopic, oxidative stress, and gene expression analyses. Cryobiology. 2020;96:19–29.
- Gilbert I, Bissonnette N, Boissonneault G, Vallee M, Robert C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction. 2007;133(6):1073–1086.
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Delivering spermatozoan RNA to the oocyte. Nature. 2004;429(6988):154–154.
- Sendler E, Johnson GD, Mao S, et al. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013;41(7):4104–4117.
- Shafeeque CM, Singh RP, Sharma SK, et al. Development of a new method for sperm RNA purification in the chicken. Anim Reprod Sci. 2014;149(3–4):259–265.
- Yang CC, Lin YS, Hsu CC, Wu SC, Lin EC, Cheng WTK. Identification and sequencing of remnant messenger RNAs found in domestic swine (Sus scrofa) fresh ejaculated spermatozoa. Anim Reprod Sci. 2009;113(1–4):143–155.
- Pang WK, Kang S, Ryu DY, Rahman MS, Park YJ, Pang MG. Optimization of sperm RNA processing for developmental research. Sci Rep. 2020;10(1):11606.
- Tiwari A, Singh D, Kumar OS, Sharma MK. Expression of cytochrome P450 aromatase transcripts in buffalo (Bubalus bubalis)-ejaculated spermatozoa and its relationship with sperm motility. Domest Anim Endocrinol. 2008;34(3):238–249.
- Savadi‐Shiraz E, Edalatkhah H, Talebi S, et al. Quantification of sperm specific mRNA transcripts (PRM1, PRM2, and TNP2) in teratozoospermia and normozoospermia: New correlations between mRNA content and morphology of sperm. Mol Reprod Dev. 2015;82(1):26–35.
- Boerke A, Dieleman SJ, Gadella BM. A possible role for sperm RNA in early embryo development. Theriogenology. 2007;68(Suppl 1):S147–S155.
- Arangasamy A, Kasimanickam VR, DeJarnette JM, Kasimanickam RK. Association of CRISP2, CCT8, PEBP1 mRNA abundance in sperm and sire conception rate in Holstein bulls. Theriogenology. 2011;76(3):570–577.
- Kasimanickam VR, Kasimanickam RK, Kastelic JP, Stevenson JS. Associations of adiponectin and fertility estimates in Holstein bulls. Theriogenology. 2013;79(5):766–777.
- Das PJ, McCarthy F, Vishnoi M, et al. Stallion sperm transcriptome comprises functionally coherent coding and regulatory RNAs as revealed by microarray analysis and RNA-seq. PLOS One. 2013;8(2):e56535.
- Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. The sperm nucleus: chromatin, RNA and the nuclear matrix. Reproduction. 2011;141(1):21–36.
- Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Arch Androl. 2007;53(3):161–167.
- Georgiadis AP, Kishore A, Zorrilla M, et al. High quality RNA in semen and sperm: isolation, analysis and potential application in clinical testing. J Urol. 2015;193(1):352–359.
- Lalancette C, Thibault C, Bachand I, Caron N, Bissonnette N. Transcriptome analysis of bull semen with extreme nonreturn rate: use of suppression-subtractive hybridization to identify functional markers for fertility. Biol Reprod. 2008;78(4):618–635.
- Bissonnette N, Levesque-Sergerie JP, Thibault C, Boissonneault G. Spermatozoal transcriptome profiling for bull sperm motility: a potential tool to evaluate semen quality. Reproduction. 2009;138(1):65–80.
- Hwang JY, Mulligan BP, Kim HM, Yang BC, Lee CK. Quantitative analysis of sperm mRNA in the pig: relationship with early embryo development and capacitation. Reprod Fertil Dev. 2013;25(5):807–817.
- Varner DD, Johnson L. From a sperm’s eye view: revisiting our perception of this intriguing cell. In AAEP Proceedings. 2007;53:104–177.
- Zhao Y, Li Q, Yao C, et al. Characterization and quantification of mRNA transcripts in ejaculated spermatozoa of fertile men by serial analysis of gene expression. Hum Reprod. 2006;21(6):1583–1590.
- Vasisth R, Gurao A, Kumari N, et al. Development and validation of most efficient RNA isolation method from buffalo bull spermatozoa. Mol Biol Rep. 2023;50(8):6717–6727. pp.
- Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15(3):155.
- Arias ME, Andara K, Briones E, Felmer R. Bovine sperm separation by Swim-up and density gradients (Percoll and BoviPure): Effect on sperm quality, function and gene expression. Reprod Biol. 2017;17(2):126–132.
- Platts AE, Dix DJ, Chemes HE, et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16(7):763–773.
- Bianchi E, Stermer A, Boekelheide K, et al. High‐quality human and rat spermatozoal RNA isolation for functional genomic studies. Andrology. 2018;6(2):374–383.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159.
- Parthipan S, Selvaraju S, Somashekar L, Kolte AP, Arangasamy A, Ravindra JP. Spermatozoa input concentrations and RNA isolation methods on RNA yield and quality in bull (Bos taurus). Anal Biochem. 2015;482:32–39.
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet. 2003;35(4):292–293.
- Imbeaud S, Graudens E, Boulanger V, et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33(6):e56.
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual (No. Ed. 2). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989.
- Jodar M, Sendler E, Moskovtsev SI, et al. Absence of sperm RNA elements correlates with idiopathic male infertility. Sci Transl Med. 2015;7(295):295re6-295re6.
- Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet. 2016;17(12):733–743.
- Burl RB, Clough S, Sendler E, Estill M, Krawetz SA. Sperm RNA elements as markers of health. Syst Biol Reprod Med. 2018;64(1):25–38.
- Yang C, Lin Y, Hsu C, Tsai M, Wu S, Cheng W. Seasonal effect on sperm messenger RNA profile of domestic swine (Sus Scrofa). Anim Reprod Sci. 2010;119(1–2):76–84.
- Chen X, Wang Y, Zhu H, et al. Comparative transcript profiling of gene expression of fresh and frozen–thawed bull sperm. Theriogenology. 2015;83(4):504–511.
- Fraser L. Sperm transcriptome profiling for assessment of boar semen freezability. IJASRM. 2016;1(12):9–12.
- Gross N, Penagaricano F, Khatib H. Integration of whole‐genome DNA methylation data with RNA sequencing data to identify markers for bull fertility. Anim Genet. 2020;51(4):502–510.
- Somfai T, Bodo S, Nagy S, et al. Effect of swim up and Percoll treatment on viability and acrosome integrity of frozen–thawed bull spermatozoa. Reprod Domest Anim. 2002;37(5):285–290.
- Mao S, Goodrich RJ, Hauser R, Schrader SM, Chen Z, Krawetz SA. Evaluation of the effectiveness of semen storage and sperm purification methods for spermatozoa transcript profiling. Syst Biol Reprod Med. 2013;59(5):287–295.
- Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139(2):287–301.
- Sahoo B, Guttula PK, Gupta MK. Comparison of spermatozoal RNA extraction methods in goats. Anal Biochem. 2021;614:114059.
- Perumal P, Chamuah JK, Srivastava N, Vupru K, Srivastava SK. Infectious causes of infertility in buffalo bull (Bubalus bubalis). Int J Bio-Res Stress Manag. 2013;4(1):84–90.
- Ibrahim S, Mahmoud KGM, Sosa AS, Sakr AAM, El‐Naby A, Nawito MF. Establishment a protocol for total RNA isolation from buffalo fresh and frozen semen for molecular applications. Andrologia. 2020;52(4):e13526.
- Vijayalakshmy K, Kumar P, Virmani M, et al. A novel combination of silane‐coated silica colloid with hybrid RNA extraction protocol and RNA enrichment for downstream applications of spermatozoal RNA. Andrologia. 2018;50(6):e13030.
- Raval NP, Shah TM, George LB, Joshi CG. Insight into bovine (Bos indicus) spermatozoal whole transcriptome profile. Theriogenology. 2019;129:8–13.
- Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7(1):3.