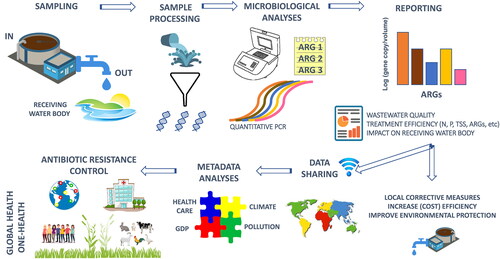
Abstract
Antibiotic resistance is a major threat to human-health and wellbeing. Antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) are environmental contaminants that circulate among humans, animals and the environment. In urban areas, wastewater treatment plants are the major recipients of these contaminants. Despite the partial elimination during treatment, final effluents, even after disinfection, contain high doses of ARB&ARG. The consequent continuous discharge of these effluents has important adverse impacts, which are particularly intense in vulnerable and deteriorated receiving environments (e.g., due to pollution, droughts or floods, reduced biodiversity). ARB&ARGs are biological contaminants capable of self-replication and horizontal gene-transfer, capabilities that due to pollution-induced selective pressure effects or absence of competition can be enhanced in deteriorated environments. Moreover, as other contaminants, ARB&ARGs can be transported, mainly through water, increasing the risks of circling back as a source of exposure to humans. The current knowledge about antibiotic resistance implications in terms of environmental contamination and risks to human-health, as well as the advances on wastewater treatment technology and antibiotic resistance quantification methods, support the need and timeliness of implementing regular wastewater monitoring systems. Because no single chemical or microbiological parameter can be used to infer the antibiotic resistance load, its specific monitoring should be part of the parameters used to assess wastewater quality. The definition of minimal requirements and integrated monitoring are essential to map antibiotic resistance at time- and space scales, and to design and implement corrective measures. These goals are technically and economically feasible and should be incorporated into wastewater quality directives.
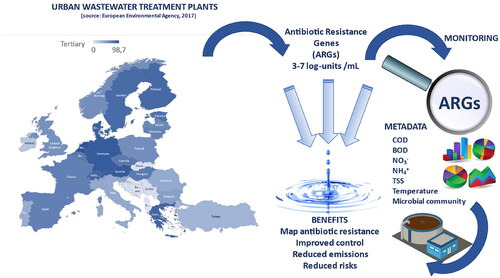
Graphical abstract
Handling Editors:
1. Introduction
In 2019, the World Health Organization included Antimicrobial Resistance as one of the ten threats to global health (World Health Organization, Citation2019). An important fraction of antimicrobial resistance is represented by ARB, mainly hosted and emitted by humans. A recent report estimated that by 2017, 55% of the global population still lacked access to safely managed sanitation (UN-Water, Citation2020). This a major threat to global health since UWTPs are essential to attenuate the impacts caused by the dissemination of ARB&ARGs and pathogens. However, it has been evidenced that UWTPs are not capable of eliminating these biological contaminants, which are continuously disseminated to the environment (Berendonk et al., Citation2015). The elimination of ARB&ARGs during wastewater treatment is influenced by a complex interplay of factors, challenging the development of improved treatment processes and leading to important differences in the quality of treated effluents (Ashbolt et al., Citation2018; Krzeminski et al., Citation2019; Pallares-Vega et al., Citation2021; Pärnänen et al., Citation2019; Rizzo et al., Citation2020). Despite the increasing number of scientific publications in the field over the last years, data about removal and discharge of antibiotic resistance by UWTPs is still scattered (Miłobedzka et al., Citation2022). Nonetheless, low removal rates and high impacts produced by UWTPs are well documented in the literature (Section 2.). This situation calls for the need of establishing regular monitoring of antibiotic resistance in the influent and final effluent of UWTPs and the recommendation of minimal removal rates. This information would contribute to measure and map the impacts of UWTPs discharges and support the decision about the treatment improvements that may be required. Finally, it would contribute to minimize the negative impacts of ARB&ARG discharges by UWTPs. A first step to meet this objective is to include in the urban wastewater treatment directive, a recommendation for ARB&ARGs monitoring and define minimal quality requirements for effluent discharging. This review aims to demonstrate that UWTPs worldwide have important impacts on the surrounding environment, contributing for the dispersal of AR and for raising the associated human health risks. Based on the current knowledge and previous experience on antibiotic resistance quantification, this review aims to propose the definition of minimum quality requirements and of integrated monitoring schemes that will contribute to halt antibiotic resistance dissemination through wastewater.
2. Conventional urban wastewater treatment partially removes ARB&ARGs
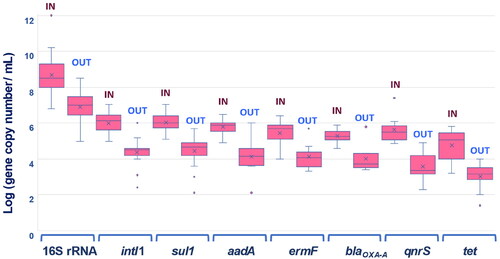
Data referring to more than 100 full-scale UWTPs operating in India, China, Singapore, Tunisia, Finland, Estonia, Germany, Netherlands, Italy, Spain, Portugal, United States of America and Canada show that raw influents have about 8–10 log-units/mL of total bacteria, measured based on the 16S rRNA gene. The reported values, determined based on qPCR, in general ranged 8–9 log-units/mL in Europe and United States of America, being slightly higher in China (9–10 log/mL) or India (12 log-units/mL, 3 UWTP, single study) (; Table SI-1) (Saxena et al., Citation2021; Wang et al., Citation2017). Also in raw influent, ARG abundance was up to 3–7 log-units/mL, according to data collected for ∼ 90 UWTPs (Table SI-1). In general, final effluents contain 6–8 log-units/mL of 16S rRNA gene, suggesting that around 2-log-units of bacteria can be removed during wastewater treatment (; Table SI-1). The difference between the bacteria or gene abundance in the final effluent and in the influent is a measure of the removal capacity of the wastewater treatment process. The 16S rRNA gene and ARG removal values reported in the literature are variable, and can hardly be explained based on the geography, size, technology, or operational conditions of the UWTP. Still, reported removal values range 1–3 log-units (per volume), being, in general, higher for ARGs than of the 16S rRNA gene (Table SI-1). Typically, the removal (per volume) observed for 16S rRNA gene and for ARGs does not differ by more than 0.2–0.5, which implies that the ratio ARGs/16S rRNA is reasonably stable over the treatment process. Therefore, it is recommended to report quantification of ARGs normalized to the sample volume, when the aim is to quantify removal rates and assess treatment efficiency. The literature reveals that regardless the specificities of each UWTP, there are common patterns and trends. Some studies can be described as examples.
Figure 1. Abundance of genes determined based on quantitative PCR in the raw influent (IN) and final effluent (OUT) of UWTPs in different countries and published in different studies. Data corresponds to determinations made in n UWTPs: 16S rRNA gene (n = 125 IN; n = 32 OUT); intI1 (n = 80 IN; n = 18 OUT); sul1 (n = 17 IN; n = 16 OUT); aadA (n = 11 IN; n = 10 OUT); ermF (n = 13 IN; n = 10 OUT); tet (n = 18 IN; n = 12 OUT); blaOXA (n = 12 IN; n = 9 OUT); qnrS (n = 7 IN; n = 6 OUT). Please see text and Table SI-1 for other details.
Pärnänen et al. (Citation2019) analyzed 12 UWTPs in distinct European countries (Southern/Western Europe - Portugal, Spain, Cyprus, Ireland and Central/Northern Europe – Germany, Finland, Norway). In Southern European UWTPs (n = 5, disregarding a non-operational UWTP due to storm overflow) were observed reduction values (log-units per volume) for 16S rRNA gene, ARGs and MGEs averaging 1.5, 1.4 and 1.3, respectively, while those values were of 2.1 in all categories for Central/Northern European countries (n = 4). The observed differences might be attributed to factors such as the size of the UWTPs (Table SI 1), higher human consumption of antibiotics or the annual temperature range. Pallares-Vega et al. (Citation2019) analyzed 62 UWTPs in The Netherlands, with different sizes and treatment processes. The highest removal values ranged 2.8–2.9 for 16S rRNA, 3.3–3.5 for ARGs, 2.4–2.6 for MGEs and the lowest 0.9 for 16S rRNA, 1.0–1.1 for ARGs, −0.1–0.5 for MGEs. Factors such as UWTP size, treatment steps or secondary treatment options had little influence on the observed removal values. The authors concluded that rainfall was the condition that most negatively affected treatment efficiency and that the presence of healthcare institutions in the served area slightly increased the concentrations of ARGs or MGEs in influent. These examples illustrate the unpredictability and complexity of factors that may influence antibiotic resistance removal during wastewater treatment. More examples are presented in Table SI-1 and a summary is provided in Box 1.
Box 1. What do we know about ARB&ARG removal during wastewater treatment?
The type of wastewater treatment process cannot per se explain the capacity to remove ARGs;
The current knowledge suggests that operational conditions and external variables (e.g., load of chemical and physical contaminants in the raw influent and the treatment biomass, climate conditions) may determine the extent of ARG removal;
Bacteria, ARB or not, are removed at approximately similar rates, therefore efforts on wastewater treatment should be focused on the elimination of bacteria;
Small UWTPs may present important fluctuations of ARG input and removal, as unexpected discharges or storm overflows have considerable impacts on the functioning of the system;
UWTPs in technical/operational failure and storm overflows may contribute to increase the load of ARGs in the receiving environment;
Disinfection processes need careful operational optimization and regular monitoring to maximize the cost- effective removal of ARB&ARGs;
Even the treatment processes with the highest removal capacities reported so far produce effluents with 3–5 log-units/mL of ARGs, including emerging ARGs.
3. UWTPs ARB&ARGs emissions and impacts
3.1. Impacts: a compromise between emissions and receiving environment
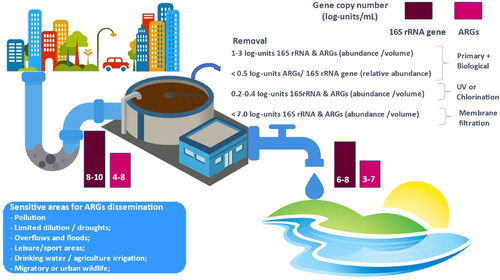
Regardless the type of treatment implemented, UWTPs may discharge into the receiving environment effluents that contain ARGs up to 7 log-units/mL (Table SI-1). River and lake water is reported to contain about 3–7 log-units/mL of bacteria (16S rRNA gene), which implies that ARGs emissions may have strong impacts on the autochthonous microbial communities (Harnisz et al., Citation2020; LaPara et al., Citation2011; McConnell et al., Citation2018a). These continuous emissions correspond to values of 13–17 log-units/day of ARGs discharged by small UWTPs (10 000 m3/day) and of 15–19 log-units/day of ARGs discharged by large UWTPs (1 000 000 m3/day). Depending on the dilution and attenuation capacity of the receiving environment, these emissions may have variable effects, severely aggravated under water stress scenarios (Keller et al., Citation2014). The impacts of UWTPs on the receiving environment (rivers, creeks, lakes or coastal areas) have been reported worldwide.
As the literature shows, these impacts can cause increases of ARG abundance and diversity, spreading over variable distances downstream of the discharge point or accumulating in sediments or biofilms. The contamination of the downstream rivers by ARGs due to UWTPs discharges is demonstrated in multiple studies. For instance, Quintela-Baluja et al. (Citation2019) reported a dramatic increase in the diversity of ARGs in the downstream river water (140 ARGs) and sediments (122 ARGs), compared with the upstream sampled sites (80 ARGs). Raza et al. (Citation2021) observed that the relative abundance of 15 ARGs, assessed based on metagenomics, increased significantly in the river in South Korea, downstream of the discharge point of each of 8 UWTPs. Also, multiple studies conducted in Catalonia (Spain) showed similar effects of increases of ARG abundance and diversity downstream of the UWTPs discharges (Lekunberri et al., Citation2017; Citation2018; Marti et al., Citation2013; Rodriguez-Mozaz et al., Citation2015). The cumulative effects of ARGs discharges have been also suggested. Quintela-Baluja et al. (Citation2019) showed that that the highest concentration of ARGs, transposon and integrase genes was observed downstream, in river sediments. These increases can reach to 2 log-units/g of ARGs in river sediments downstream of the UWTPs discharge points have been reported (Brown et al., Citation2019; Quintela-Baluja et al., Citation2019). These results suggest that sediments may represent important reservoirs where ARGs emitted by UWTPs are deposited over time. Also, biofilms may be important ARG reservoirs. Cacace et al. (Citation2019) used qPCR to compare the final effluent of 12 UWTPs and biofilms collected in the receiving rivers in 8 different European countries (France, Norway, Portugal, Germany, Netherlands, Turkey, Austria and United Kingdom). The ratio between the abundance of ARG downstream of the effluent discharge site and the corresponding upstream value, was >1 in most of the analyzed situations, suggesting a measurable impact of UWTPs (Cacace et al., Citation2019). Other authors have also called the attention for the fact that also MGEs can present increased abundance downstream of the UWTPs discharging point, suggesting the potential to enhance ARG dissemination through horizontal gene transfer (Akiyama et al., Citation2010; Lekunberri et al., Citation2017). Transport is another mechanism of enhancing AR dissemination. This effect was shown by Thornton et al. (Citation2020), who demonstrated that ARGs were more abundant and diverse up to 5 km downstream the point of discharge of a UWTP into the Blue river in Colorado (USA). Similar observations were reported by LaPara et al. (Citation2011), who highlighted the fact that such impacts could not be avoided through the implementation of tertiary treatment.
Given the comparatively lower flow rates, lakes may be even more vulnerable than rivers to the impacts caused by UWTPs. In a study examining 21 lakes impacted by distinct contamination sources, in Switzerland, Czekalski et al. (Citation2015) explored the relationship between human activities, microbial community composition and the eutrophication status. The authors concluded that the observed effects increased with the number and capacity of UWTPs in the catchment as well as with the levels of eutrophication. The potential role of sediments on the accumulation of resistance determinants was also demonstrated in lakes. Devarajan et al. (Citation2015) explored the effects of changing in 2001 of the point of discharge of a UWTP operating since 1964 and emitting the final effluents into the Lake Geneva (Switzerland). The study showed that increased eutrophication levels were accompanied by the vertical accumulation of Escherichia coli, enterococci, and Pseudomonas in the original discharge point, while recent beta-lactamase encoding ARGs (blaCTX-M, blaSHV and blaNDM) were detected in the top layer of the sediments, close to the new discharge point. Also in lakes, contaminant ARGs can be transported in the receiving environment. Chu et al. (Citation2018) showed a significant negative correlation between ARG diversity, assessed based on metagenomic analysis, and distance, up to 5 km from the points where two UWTPs discharged into Lake Michigan (US).
Impacts are often assessed based on the most abundant ARG emitted by UWTPs, which are those most suited for source-tracking (Li et al., Citation2018). Examples of these include ARGs encoding resistance to aminoglycosides, beta-lactams, tetracycline, and macrolide-lincosamide-streptogramin B (MLSB) and multidrug-efflux pumps. Indeed, these are those producing the most noticeable impacts on the receiving environment (An et al., Citation2018; Chu et al., Citation2018; Quintela-Baluja et al., Citation2019; Thornton et al., Citation2020). Nevertheless, emerging ARGs have also been reported in environments that receive UWTPs effluents. Proia et al. (Citation2018a) reported that ARGs such as the carbapenemase encoding genes blaOXA-48, blaKPC or blaNDM were detected in 96%, 93% and 68% of the Zenner River (Belgium) samples analyzed, respectively. Although the wastewater treatment reduced the abundance of these genes (reduction of 1.9–2.6 log-units per volume), the discharge caused a general increase from upstream to downstream the discharge point in the river. Amos et al. (Citation2014) made a similar study over a three years period (2009–2011) in the sediments of Thames River (UK). The authors reported a dramatic increase of third-generation cephalosporin (3GC)-resistant Enterobacteriaceae downstream of the UWTP discharge point. The study focused on the blaCTX-M-15, harbored by the pathogenic clone Escherichia coli ST131 and presenting distinct MGEs rearrangements, suggesting the mobilization of the gene.
3.2. Impacts are stronger in vulnerable receiving environments
The impacts caused by the discharges of UWTPs may be aggravated or attenuated by factors that are external to the UWTPs (). High pollution and eutrophication levels in the receiving environment may favor the overgrowth of minor populations, mainly anthropogenic or animal associated bacteria. Floods and droughts are also important for determining the severity and extent of UWTPs impacts. Flood events have the potential to enhance UWTPs impacts, due to the release of nutrients and ARGs from UWTPs, combined with the leaching contaminants from other areas. Drought events have also the capacity to enhance UWTPs impacts, since the effluent may represent most of the water in the receiving environment, therefore with minimal dilution of ARGs, MGEs, nutrients and other contaminants. These effects have been illustrated in different studies (Kalinowska et al., Citation2021; Marti et al., Citation2013; McConnell et al., Citation2018a; Storteboom et al., Citation2010; Zhang et al., Citation2013). Storteboom et al. (Citation2010) tracked the sources of ARGs (2 sul and 11 tet genes) inputs along nine sites in the Cache la Poudre (Poudre) and South Platte Rivers in Colorado (USA), putatively impacted by UWTPs and/or by agricultural sources. The authors concluded that UWTPs might be a dominant source of ARGs to the river and argued about the synergistic effect with other pollutants that could contribute to amplify the impacts of these discharges. Indeed, the impacts caused by UWPTs on the alteration of physicochemical conditions and microbial community have been demonstrated. Kalinowska et al. (Citation2021) showed how a small-scale wastewater effluent discharge disturbed the physicochemical conditions and the microbial community in an oligotrophic lake (in Poland). The authors reported a slight increase of electric conductivity and total nitrogen content, while the microbial community mirrored that of the effluent, with the predominance of Proteobacteria (29%) and Bacteroidetes (15%). These observations are supported by a prior study by Marti et al. (Citation2013) who observed that Gammaproteobacteria, most of which presumably related with human activities and present in the UWTP effluent, considerably increased in the downstream river biofilm samples compared with upstream. Also, the concentrations of NH4+-N and PO43- have been suggested (Zhang et al., Citation2013) to be associated with increased loads of ARGs in the Jiulong River (China), either because they are discharged together and/or because those compounds act as bacterial nutrients. The relationship between nutrients, especially nitrogen species, and ARGs has been noted by other authors (Devarajan et al., Citation2015; Freeman et al., Citation2018). While the discharge of nutrients may lead to the over proliferation of some minor populations (Chu et al., Citation2018), in the presence of high nutrient loads, the bacteria discharged in the effluent tend to increase in the receiving environment (Marti et al., Citation2013). Different studies argue on the possible selective effect or simultaneous occurrence of antibiotic residues and ARB/ARGs, observed to be positively correlated in UWTPs discharging sites (Proia et al., Citation2018b; Rodriguez-Mozaz et al., Citation2015). However, others failed to find such correlations, at least between a specific antibiotic and the corresponding antibiotic resistance pheno- or genotypes (Narciso-da-Rocha et al., Citation2014; Novo et al., Citation2013; Pärnänen et al., Citation2019; Varela et al., Citation2014). Selection is further discussed in section 5.
Figure 2. Pathways of dissemination and/or removal of antibiotic resistance by urban wastewater treatment plants and potential impacts. Examples of sensitive areas that may be impacted by UWTPs are provided: areas subjected to other pollution sources (industry, intensive agriculture); areas that can experience overflow and flood events or with limited dilution capacity, particularly in drought regions and/or seasons; discharging in proximity to recreational areas or where water for drinking water production or agricultural irrigation are collected.
4. Advanced treatment for ARB&ARG removal: limited efficiency calls for case-by-case optimization
The literature reveals multiple options available or under development that include chemical (oxidation)-based processes (e.g., UV, chlorination, ozonation, peracetic acid), physical separation (filtration processes such as ultrafiltration, nanofiltration or reverse osmosis), biological processes (MBR, constructed wetlands) and soil aquifer treatment (Pei et al., Citation2019). While the efficacy of some of these processes, mostly UV, chlorination, peracetic acid and MBR has been studied at full-scale UWTPs, others are mainly explored at the laboratory scale and, less frequently, at pilot-scale (Rizzo et al., Citation2020). Pilot- and laboratory-scale assays are excellent to compare the efficiency of different processes, explore the influence of specific variables or infer about possible side- and unintended effects, however, have a limited capability of predicting the behavior of these advanced processes in full-scale UWTPs. In this review, only advanced processes implemented in full-scale UWTPs are discussed.
Chlorination shows a limited capacity to reduce the ARB&ARGs, as has been extensively reviewed (Hiller et al., Citation2019; Liu et al., Citation2018; Pei et al., Citation2019; Rizzo et al., Citation2020). Thakali et al. (Citation2020) reported the use of chlorination in two UWTPs (USA) with an aeration basin/final settling tank/chlorination (277 000 m3/day) and an aerobic digester/aerobic basin/clarifier/chlorination (60 000 m3/day), respectively. ARG reduction values (log-units/mL) due to chlorination ranged 0.4 ± 0.5. Also, very low or nil removal rates were reported by Oliveira et al. (Citation2020) who investigated two UWTPs (Portugal) with biological aerated filters (756 000 p.e.) or conventional activated sludge and sand filtration (211 000 p.e.), both with sodium hypochlorite disinfection. Chlorination led to reduction values (+0.4 or − 0.3) that suggested a negligible or even adverse effect of the disinfection step. Liu et al. (Citation2018) investigated the role of chlorination in the variation of ARGs collected from intracellular fractions (iARGs) and free-DNA (eARGs). Compared to the secondary effluent, iARGs relative abundance (per 16S rRNA gene) after chlorination presented increases up to 25.5 fold (ARG qnrA, quinolone resistance) or up to73.3 (ARG vanA, vancomycin resistance), while others suffered very low reduction values. According to these authors, also eARGs increased up to 621.2 fold compared with untreated wastewater (vanA). Yi et al. (Citation2015) measured the removal of beta-lactamase encoding genes after chlorination in four UWTPs and concluded that the genes blaCTX-M, blaOXA and blaTEM presented reductions of about 80% (normalized per dry influent biomass), while blaSHV increased in the four UWTPs up to 500%. An alternative to chlorination is peracetic acid. Fiorentino et al. (Citation2019) examined the performance of this disinfection process, using chemical treatment (aluminum polychloride enriched by sodium hydroxide), and a final disinfection step by peracetic acid. The authors concluded that disinfection by peracetic acid was not effective to remove ARGs and intI1, with log reduction values (per volume) close to zero.
Also for UV, the literature suggests an almost negligible effect on ARG removal. Wen et al. (Citation2016) reported ARGs reduction values of 0.20–0.33 (log-units/mL) due to UV disinfection (fluence not reported), which agrees with other studies. Narciso-da-Rocha et al. (Citation2018) observed that UV (29.7 mJ/cm2) had little effect on ARBs, with reductions of 1–28% and had no effect on the examined ARGs, causing a significant increase of the gene intI1. However, post-UV water storage was associated with a significant decrease of some ARGs, although with the increase of Betaproteobacteria and Flavobacteria. Yang et al. (Citation2019a) registered less than 0.4 reduction values (log-units/mL) of ARGs and 16S rRNA gene after UV disinfection (20 mJ/cm2). Yu et al.,(Citation2020) studied the effect of UV disinfection (<30 mJ/cm2) in two UWTPs and compared the efficiency of disinfection on particles-associated and cell-free DNA. They observed that the relative abundance of ARGs and MGEs increased in the particles-associated fraction and slightly decreased in the cell-free fraction after UV disinfection, suggesting that UV radiation might be quenched by particles. However, coagulation and filtration may be not enough to avoid such interference effect. When coagulation and filtration were applied before UV disinfection (27 mJ/cm2), ARGs were not reduced, and ARB presented small reduction values (34–75%) (Lee et al., Citation2017). Jia et al. (Citation2021) studied the effect of UV disinfection (20 mJ/cm2) in a UWTP and observed an increase in the relative abundance of ARGs conferring resistance to aminoglycoside, tetracycline, sulfonamide, chloramphenicol, and macrolide-lincosamide-streptogramin B and a decrease of ARGs encoding bacitracin and beta-lactam resistance. Lin et al. (Citation2021) observed that UV performed better than chlorination on the reduction of the relative abundance (per 16S rRNA gene) of indicator genes (99.99% and 97.2% vs. 51.6 and 85.6% for intI1 and intI2, with UV and chlorination, respectively). Chen and Zhang (Citation2013) compared three UWTPs using constructed wetland (including four stabilization ponds and one horizontal subsurface flow wetland), aerated biological filter (1500 m2 with a 3 h HRT) and UV (45% transmittance, total power of 900 kW, and light intensity > 1 mW/mm2), as post-secondary treatment. The results ranked these processes as constructed wetland > aerated biological filter > UV, with ARG reduction values (log-unit per volume) of 1.3–2.1, 1.0–1.21 and 0.5–0.7, respectively. One of the impairments of UV disinfection might be solved with the use of adequate radiation dosage. McConnell et al. (Citation2018b) compared doses of 50 and 250 mJ/cm2 and observed significant differences on ARG removal.
Disinfection processes such as those mentioned above have the potential to interfere with cell structures with different implications: i) lead to the leak of the cell content, and cause the release of eARGs (Liu et al., Citation2018); ii) trigger stress responses that may increase the bacterial resilience to elimination or ARGs transfer potential (Di Cesare et al., Citation2016; Narciso-da-Rocha et al., Citation2018); iii) activate repair and survival mechanisms, particularly efficient is some bacterial groups, permitting regrowth after the relief of the stress conditions imposed by advanced treatment, which may lead to ARG increase and/or microbial community disturbances (Becerra-Castro et al., Citation2016; Sousa et al., Citation2017). On the other side, the efficacy of disinfection processes can be jeopardized by the presence of particles, organic matter, dead cells, other type of debris, among other. Such side-effects and impediments may be situation-specific, but globally show the limited capacity of existing disinfection processes to effectively reduce ARGs, highlighting the unpredictability of challenges occurring under field conditions. A key message refers to the need to optimize and regularly monitor and adjust the operational conditions of disinfection installed in UWTPs.
Membrane bioreactor (MBR) systems have been suggested as potential successful alternatives or complements to conventional wastewater treatment processes. Lin et al. (Citation2021) reported a reduction of ARGs of 3.5 log-units/mL in a UWTP (China, size not provided) using A2O process complemented by MBR. This was an improvement, when compared with a parallel line where a high efficiency flocculent settling/cloth media filter (HEFS/CMF) complemented the A2O, with reduction values of 2.1 log unit/mL. Du et al. (Citation2015), studying a small scale UWTP (China, 30 000 m3/day and with 50% industrial effluents) with A2O/MBR (hydrophilic polyvinylidene fluoride hollow fiber membrane with mean pore size of 0.1–0.4 μm) reported reduction values that ranged 0.7 to 4.7 log-units/mL. The authors highlighted the high efficiency of MBR to remove ARGs from wastewater. Le et al. (Citation2018) compared the capacity of a line of a UWTP (Singapore, size not provided) that combined anoxic-aerobic treatment and second clarifier with another that used microfiltration/MBR. The microfiltration treatment was observed to reduce ARB to levels below the detection limit, the 16S rRNA gene in the range of 1.6–3.6 and ARGs up to 7.1 (log-units/mL) in the whole MBR train. However, importantly, the authors noted that genes such as blaKPC, blaNDM, blaSHV, ermB, intI1, sul1, and tetO were still detected at average concentrations up to 2 log-units/mL in the final effluent, possibly due to persistence of free DNA (Le et al., Citation2018). Indeed, it has been argued that MBR effluents may still contain high loads of ARGs (∼3–5 log-units/mL) (Lin et al., Citation2021; Zheng et al., Citation2019). Moreover, these effluents may hold disturbed microbial communities, dominated by members of genera such as Pseudomonas, Acinetobacter, Varivorax, Comamonas, Thermomonas, Acidovorax, Delftia, Sphingomonas, Chryseobacterium, Azospira, and Flavobacteria (typical of human-intervened or impacted water) (Ng et al., Citation2019).
5. The known and unknown factors that drive antibiotic resistance dissemination
Acquired AR involves two major mechanisms, the ARG acquisition, through horizontal gene transfer (mostly transformation, transduction, conjugation) or mutation. While the first is more relevant for the spread of new genes, the second is an important mechanism of evolution of novel allelic variants (Davies & Davies, Citation2010). Depending on the bacterial host and environmental context, in a population, acquired AR may be readily eliminated, persist at stable levels, or over proliferate. In the latter case, this is due to any kind of selective advantage, through which a specific clone, lineage, or sub-population multiplies and amplifies the acquired ARGs by vertical gene transfer (Davies & Davies, Citation2010). Irrespective of the moment when acquisition occurs, it is the selective advantage of the clone or lineage to survive and multiply that will determine its real impact. This capacity is a measure of the fitness of the organism, i.e., its ability to survive and multiply in a competitive environment where it can overcome conditions potentially adverse for competitors. Although some organisms have intrinsic features that permit the adaptation and proliferative success in a wide variety of environments, their fitness is always influenced by the specific conditions where the organism is thriving (). Organisms with high fitness features can behave as invasive species in a wide diversity of environments. The bacterial groups recognized among the most prominent ARB (e.g., Enterobacteriaceae, Pseudomonas, Aeromonas, Acinetobacter) have such features and can be categorized as invasive species, i.e., with the capacity to colonize, proliferate and persist in a wide array of environments, including through the urban water cycle (Vaz-Moreira et al., Citation2014). The apparent high fitness of an organism results from the combination of intrinsic and exogenous factors, selective pressures and stressors ().
Table 1. Examples of intrinsic and exogenous factors (fitness drivers) that may influence the success of an ARB in wastewater or related environment.
5.1. Synergistic or antagonistic effects of potential selectors
Among the possible selective pressures, antibiotics and metals have received much attention. Possible selective concentrations of antibiotics have been inferred based on statistical or bacterial models (e.g., Gullberg et al., Citation2011; Murray et al., Citation2021; Tello et al., Citation2012). For instance, Bengtsson-palme and Larsson (Citation2016) determined the Predicted No Effect Concentrations (PNECs) as a threshold concentration, above which an antibiotic could exert a selective pressure, meaning for example that the proliferation of non-sensitive cells can be favored in comparison with the sensitive counterparts. The PNEC values found by these authors ranged 8 to 64 000 ng/L. In general, these concentrations are below those measured in UWTPs, although values above these thresholds can be found (Rodriguez-Mozaz et al., Citation2020). However, the existing models have limited capacity to mimic complex environments and to predict the fate of ARB&ARGs during wastewater treatment. Metals and biocides have also been suggested as potential selectors of AR, mainly because both types of genes (encoding resistance to biocides or metal vs. antibiotics) may be genetically linked, facilitating the co-selection of both (Bengtsson-Palme et al., Citation2018; Gaze et al., Citation2011). For instance, Li et al. (Citation2017) observed a wide spectrum of ARGs linked to metal resistance genes that could be co-transferred by horizontal gene transfer (e.g., bacitracin and Cu/Zn/Al/Fe/Cr/As/Hg/Te; aminoglycoside and Ni/Fe/Zn; tetracycline and Zn/Fe; or beta-lactam and As/Zn/Hg.) Co-selection may also be due to the use of common resistance mechanisms against antibiotics, metals and/or biocides, for example, using efflux systems that can excrete all those types of substrate (Tong et al., Citation2021; Webber & Piddock, Citation2003). The role of biocides as possible environmental AR selectors, although if scantly investigated, has been discussed and evidenced (Paul et al., Citation2019). Oxidative stress, induced by metals, pharmaceutical compounds or nanoparticles, is also reported as being capable of promoting ARG acquisition through horizontal gene transfer (Guerin et al., Citation2009; Qiu et al., Citation2012; Wang et al., Citation2019; Zhang et al., Citation2018). Zhang et al. (Citation2018) showed that oxidative stress and SOS response were part of the effects triggered by environmentally-relevant and sub-inhibitory concentrations of Cu(II), Ag(I), Cr(VI), or Zn(II) capable of enhancing the transfer of ARGs in Escherichia coli strains through conjugation. Indeed, a recent review (Liu et al., Citation2020) reports the potential of a wide array of substances (e.g., aluminum oxide-, copper, or silver-nanoparticles, polycyclic aromatic hydrocarbons, triclosan, chloramine, hydrogen peroxide, preservatives, ionic liquids, carbamazepine, gentamicin or CO2) to promote horizontal gene transfer. However, the same review referred to a long list of other substances able to inhibit horizontal gene transfer. Antagonistic effects between metals and antibiotic selection have also been reported. For example, zinc has been suggested as capable of raising the ciprofloxacin selective concentration (Vos et al., Citation2020). These observations show how complex can be the prediction of conditions that will promote or avoid AR selection.
In addition, it is necessary to consider the complex nature of the matrix (e.g., wastewater, freshwater, sediments) and the micrometre scale environment of the bacterial cell. For instance, the biosolids onto which bacteria can adsorb may favor ARG concentration and eventual mobilization (Proia et al., Citation2018b). The potential of microplastics to simultaneously adsorb chemical pollutants and ARB and enhance the effects of selective pressures has been observed in terrestrial and aquatic environments including in UWTPs (e.g., anoxic stages, primary settling tank, selector tank, aerated tank) (Liu et al., Citation2021). Again, these examples show how difficult may be the prediction of AR selection in environments as complex as sewage and wastewater. This is probably the reason why it has been concluded that no specific selective pressures are needed to observe increases of some ARB or ARGs. For example, Lehmann et al. (Citation2016) showed that nutrients and/or microbiota supplied by a UWTPs effluent might explain the increase of the gene intI1 in river water and biofilms. In addition, it was shown that the same group of selective agents can produce an effect in a specific context and not in another. Klümper et al. (Citation2019) demonstrated that the minimal selective concentration of an antibiotic may be increased when the target bacteria are integrated in a complex microbial community. This is important evidence of the role of robust microbial communities to serve as buffer systems capable of halting ARB&ARG propagation (Ribeirinho-Soares et al., Citation2021). Exploring the drivers of ARB/ARG selection and dissemination is among the most ambitious goals in this field, yet with a long way of research to go.
5.2. AR dissemination: much more than the effect of selective pressures
A major question that has been addressed in the literature refers to the factors that might be controlled to improve wastewater treatment efficiency, as well as to reduce the impacts of final effluents. Different studies (Laht et al., Citation2014; Pärnänen et al., Citation2019) suggested that, compared with larger UWTPs, small facilities may be more vulnerable to changes in inflowing wastewater composition and flow rates. In addition, a higher daily flow may be associated with a lower hydraulic residence time, with longer periods favoring increasing ARB/ARG removal (Novo & Manaia, Citation2010; Pallares-Vega et al., Citation2021; Vilanova et al., Citation2004). Numerous studies (Bengtsson-Palme et al., Citation2016; Du et al., Citation2015; Ding et al., Citation2020; McConnell et al., Citation2018b; Novo et al., Citation2013; Tong et al., Citation2019) have identified factors such as the wastewater microbial community composition, conductivity, nutrients (nitrite, nitrate, total nitrogen, phosphate), redox potential, COD, pH, temperature, and oxygen availability to be significantly correlated with ARGs. These studies highlighted the importance of achieving a good reduction of microbial biomass and nutrients (mainly nitrogen compounds) that are supposed to be associated with the fate of ARGs during wastewater treatment. However, the stochasticity of the wastewater treatment process, seems to the major explanation for the deep variations in ARB/ARG composition observed in treated when compared with raw wastewater (Bengtsson-Palme et al., Citation2016; Lira et al., Citation2020). This evidence hints an inevitable limitation to predict the removal rate of ARB&ARGs with accuracy. The apparent randomness is, on one hand, the result of the complex interplay of variables that affect ARB&ARG survival and proliferation and, on the other side, a major limitation to fully understand the factors that may need to be controlled to maximize resistance removal. The same uncertainty rules the impacts on the receiving environments. Some studies have reported significant correlations between antibiotic residues, or metals and ARGs or ARB in rivers impacted by UWTPs (Hubeny et al., Citation2021; Proia et al., Citation2018b). Yang et al. (Citation2019b) in a study in natural wetlands observed that ammonium content in soil was positively correlated with ARGs and concluded that temperature was a determinant of ARG accumulation in riverine, wetlands, and lacustrine in the Qinghai-Tibetan Plateau. The influence of temperature, noted also by others in distinct contexts (McGough et al., Citation2020), calls the attention for potential alterations in ARG behavior due to the warming climate trend. The scenario of climate change has also increased the occurrence of floods that may cause the failure of UWTPs and contribute to increase the emission from multiple pollution sources and to leachate microcontaminants with selective pressure potential. Garner et al. (Citation2016) evaluated the abundance of ARGs in the Colorado River after an overflow and observed that although it decreased after the event, 10 months later the levels returned to values close to those observed previously, with the aggravating effect that the contamination started to spread to previously pristine areas. Also, the production of drinking water can be a problem. Despite the drinking water catchment is normally made upstream the UWTPs discharging points, the risks that microcontaminants (antibiotics, metals, among others) released in rivers and lakes will contaminate the drinking water distribution systems and exert relevant AR selection deserves attention (Rilstone et al., Citation2021). Amos et al. (Citation2015) developed three models in which the gene intI1 was used as AR indicator. As the authors showed, factors to consider in the assessment of impacts caused by UWTPs include different variables, such as the size of the plant, type of treatment, pollution and dilution effects, as well as the combination of the different water parameters. These models may be difficult to implement in complex microbial ecosystems, of which only a very reduced number of parameters is known and based on casuistic determinations. However, online monitoring, metadata and artificial intelligence tools may improve this situation in the future, permitting reliable predictive models and associated control systems (Fan et al., Citation2022).
6. Risk assessment and prevention require systematic and comprehensive data
The risks associated with ARB&ARG emitted by UWTPs can be regarded at two levels: i) the dissemination of contaminant AR in the environment, meaning direct input and potential for horizontal gene transfer and self-replication; and ii) the transmission to humans, due to higher exposure and occurrence of hazardous ARB&ARGs in the environment. Both are connected, as dissemination of ARB&ARGs leads to higher human exposure and consequently to a higher probability of transmission to humans. This is the principle that underlies the One-Health concept, which assumes that any niche where ARB can persist or develop is a potential source of transmission to humans (Hernando-Amado et al., Citation2019; McEwen & Collignon, Citation2018). ARB are mainly opportunistic pathogens, and the host colonization may be never noticed until an ARB infection is revealed. This situation may be unlikely in a healthy individual (although there are unformal reports of skin infections due to environmental ARB in healthy individuals). Therefore, ARB hosts are mostly asymptomatic carriers (colonized but not infected) who, through sewage, contribute to the dissemination of these contaminants, largely through UWTPs.
UWTPs emit final effluents with high loads of ARGs (∼3–7 log-units/mL) embedded in a complex matrix of other contaminants and nutrients, contributing to enrich the environmental ARB&ARG pool. These emissions will be directly dispersed and sometime self-replicated in the environment and, simultaneously can be infiltrated in soil/and groundwater and be transported over long distances, mainly through water and wildlife. Eventually, autochthonous environmental bacteria may also contribute to the acquisition of new ARGs. Such a scenario implies that ARB&ARGs are maximally reduced in final UWTPs effluents, although the definition of thresholds may be challenging, given the current knowledge gaps (Manaia, Citation2017). Some attempts of estimating risks of due to ARB or ARGs have been proposed in the literature. Mughini-Gras et al. (Citation2019) identified 1220 occurrences of Escherichia coli containing beta-lactamase encoding ARGs in humans (478 in patients, 742 healthy people, including poultry and pig farmers, and travelers) and 6275 occurrences in non-human sources (479 in companion animals, 4026 in farm animals, 66 in wild birds, 1308 in meat products, 51 in raw vegetables, 71 in seafood, 274 in surface freshwater). Attributed transmission from non-human sources could be ranked as food products with a mean value of 18·9% − 6.6% seafood, 1.1% raw vegetables and 11.2% meet/meet products, 2.3% for swimming in surface freshwater and 0.3% for contact with wild birds. These values were probably biased by the number of occurrences that were registered for each type of sources and therefore may be underestimated for raw vegetables, water and wildlife. Leonard, Yin, et al. (Citation2018) also measured the exposure of people practicing aquatic sports to E. coli containing beta-lactamase-encoding-ARGs. Based on the occurrence of E. coli in bathing water, the number of sport events and of practitioners and the average number of ARGs in an ARB cell (1.24), the authors estimated that in 2016 all water sports sessions in England (∼ 123 millions) might have resulted in the ingestion of one or more E. coli ARGs. Moreover, the authors estimated that 2.5 (out of ∼123) million of those sessions involved the ingestion of 100 or more ARGs harbored by E. coli. As a confirmation, the authors conducted an epidemiological survey, where they concluded that 6.3% of surfers (9/143) were colonized by E. coli containing a beta-lactamase encoding ARG, while these were only observed in 1.5% of non-surfers (Leonard, Zhang, et al., Citation2018). These results suggested that transmission from water is likely to occur, with treated wastewater being a relevant source of this form of pollution.
Nonetheless, these models may be underestimating the real impacts. The fact that are based on E. coli, which is an indicator of fecal contamination, implies an important underestimation since its presence above a defined threshold is not permitted in drinking and bathing waters or food products. Indeed, it is expected that members of this bacterial species are in lower abundance in important human-exposure sites than other ubiquitous, although relevant ARB, such as other Enterobacteriaceae (e.g., Enterobacter, Citrobacter, Klebsiella), Pseudomonas spp., Acinetobacter spp., Aeromonas spp. among other. The second limitation is that only direct transmission is considered in those studies and that is probably not the major path of transmission, since ARB&ARGs are known to spread in the environment. Therefore, these models do not consider that ARB can have intermediate hosts/habitats, for instance colonize the food products, via irrigation water influenced by wastewater, and access humans by multiple paths, in particular leisure and directly or indirectly through food-chain.
Another aspect that needs a critical evaluation refers to the limits of detection and of quantification of ARGs that can be considered high, mainly in relation to the associated risks. The major limiting factor on the establishment of the limit of quantification is the DNA extraction in which the total DNA of the sample can be concentrated 1000–5000 fold (100 − 500 mL of water/wastewater filtered result in 0.1 mL of DNA extract). Consequently, ARGs that are at abundance values ≤1 log-units/mL will not be detected in a sample. Nevertheless, this apparently low load may represent a risk, mainly if it refers to emerging ARGs that typically occur at very low abundance, until are spread and stabilized in the natural environment. A non-detected emission of rare ARGs may correspond to daily emissions of 11–12 log-units by a small UWTP (10 000 m3/day) and of 13–14 log-units in a large UWTP (1 000 000 m3/day). Probably, the effects of these emissions may only be noticed several months later, for instance in sediments or after being transported to other areas. However, these emissions are not innocuous. As self-replicative entities, ARB will be able to multiply whenever adequate conditions are met (mainly nutrients, temperature). This can occur in the receiving environment, mainly if it is already strongly impacted by humans, or in a further environment, for instance agriculture field or wildlife. Although the doubling time of bacteria is expected to be delayed in the natural environment, when compared with the ideal experimental conditions, a few days may be sufficient to reach ARB loads that are hazardous, for both types of risk – dissemination in the environment and transmission to humans. For example, considering the doubling times (hours) of 15 for E. coli, 2.3 for P. aeruginosa, 1.87 for S. aureus and 1.1 for V. cholerae (Gibson et al., Citation2018) it is possible to estimate that, in under favorable conditions and in absence of competition and predation, only two weeks may be sufficient for these bacteria reach loads ranging 8.6–25.6 log-units. These reasons contribute to explain why direct evidence for transmission to humans through the environment have not been gathered so far (Huijbers et al., Citation2015; Munck et al., Citation2015).
The types of risks posed by ARB&ARG emissions by UWTPs vary according to multiple factors that include the geographic and climate conditions, pollution load, or socioeconomic factors. Although the impacts due to UWTPs need to be minimized worldwide, some critical areas need special attention. In general, these may be the same that are signalized as sensitive to other forms of pollution and impacts, as ARB&ARGs are closely linked to chemical pollution and high loads of organic nutrients, typical of eutrophic systems. Major criteria to define sensitive or priority areas regarding ARB&ARGs include different aspects related with the capacity of ARB/ARGs to persist and proliferate and the degree of exposure, direct or indirect, of humans to these environments (, Box 2).
Box 2. Major criteria to define sensitive or priority areas regarding ARB&ARG dissemination
Receiving environments under other pollution impacts (e.g., industry, intensive agriculture), including diffuse pollution;
Limited dilution capacity of the receiving environment, particularly in drought regions and/or seasons;
Regions subjected to successive overflows and floods;
Discharging sites with direct human exposure (e.g., river or sea beaches, leisure, sports areas);
Discharging sites with indirect human exposure (e.g., drinking water catchment, agriculture irrigation);
Refuge areas of migratory or urban wildlife;
7. Monitoring antibiotic resistance
7.1. Why and where
Humans and animals that can benefit from antibiotic use are those endangered by the dissemination of ARB&ARGs as environmental contaminants. Although no evidence exists that a direct transmission of ARB may occur from wastewater to humans, the risks posed by these bio-contaminants should not be ignored (Berendonk et al., Citation2015; Manaia, Citation2017; Huijbers et al., Citation2015.). A first step to reduce the continuous emissions of contaminant ARB&ARGs by UWTPs to the surrounding environment is the implementation of regular monitoring and the definition of treatment objectives. This same strategy was used in Europe in the past for other contaminants emitted by UWTPs (e.g., N, P, organic matter) with the adoption of the Urban Waste Water Treatment Directive in 1991 (Council Directive, Citation1991, 91/271/EEC). The establishment of these criteria was an important turning point that promoted the improvement of urban wastewater treatment across the European Union, with clear gradual upgrades since 1990 to 2017 (ea.europa.eu/data-and-maps/indicators/urban-waste-water-treatment/urban-waste-water-treatment-assessment-5). The range of contaminants that today threaten the quality of effluents has been expanding over the years and AR is certainly part of this set. The inclusion of AR monitoring in the Urban Wastewater Directive would contribute to the improvement of existing treatment processes and to minimize the impacts on the surrounding environment as well as risks due to dissemination. Beside permitting the estimation of AR removal rates during wastewater treatment, monitoring should focus also on impacts, since an effluent even with a low AR load may have a negative impact on a sensitive or fragile receiving environment (Box 2). Moreover, because ARB&ARGs can accumulate in the receiving water body and in sediments, long-term impacts should be assessed on a regular basis. Hence, corrective measures could include interventions in the treatment process and/or at the receiving environment, aiming at reducing the impacts of UWTPs emissions. Since ARB&ARGs have been observed to be correlated with multiple factors, AR monitoring could be complemented by demographic (big)data and geographic information systems. The monitoring of UWTPs emissions and of the receiving environments is essential to identify critical points of AR discharge and accumulation and to produce integrated pollution maps, associated with UWTPs. These systems might be articulated with other One-Health surveillance schemes.
7.2. Targets and methods
Based on the current knowledge, as summarized in section 5, it is not possible to predict AR levels from any kind of surrogate data (e.g., antibiotic residues, nutrients). Therefore, AR must be monitored directly, i.e., ARB or ARGs. Among ARB, E. coli is a likely candidate to be monitored, used for example in the Global Tricycle Surveillance (World Health Organization, Citation2021) and in different studies (Ferreira da Silva et al., Citation2007; Honda et al., Citation2020; Marano et al., Citation2020; Novo & Manaia, Citation2010; Pallares-Vega et al., Citation2021). However, in wastewater and water most ARGs hosts are non-culturable and/or non-enteric bacteria, such as Pseudomonas, Acinetobacter, Aeromonas, among other (Dai et al., Citation2022; Narciso-da-Rocha et al., Citation2018). Moreover, the same gene may be harbored by members of different taxa (Keenum et al., Citation2022). E. coli and related bacteria (coliforms) have been used as microbiological indicators of fecal contamination, often interpreted as a measure of human health risk, although fail to properly represent waterborne pathogens (Korajkic et al., Citation2018). Although E. coli is still a reference to define threshold values to categorize water quality and safety (Table SI-2), it is not an adequate AR indicator in wastewater. A recent study that analyzed 228 influent and 244 effluent samples of 57 UWTPs located in 22 countries of five continents showed that coliforms abundance, in which group E. coli was the predominant species, ranged 5.5 log-units of CFU/mL in the influent and 3.0 log-units of CFU/mL in the effluent (Marano et al., Citation2020). Different studies have shown that wastewater enterobacteria, including E. coli, may yield antibiotic resistance percentage values that vary between <1% to >20%, against penicillins, cephalosporins, quinolones, tetracycline, among other (Honda et al., Citation2020; Hutinel et al., Citation2019; Marano et al., Citation2020; Narciso-da-Rocha et al., Citation2018). These observations suggest that the measurement of antibiotic resistant E. coli might imply the need to analyze large volumes of water, and multiple culture media (supplemented with antibiotics). Despite the importance of this microbiological indicator, transecting different type of environments, it represents a minor fraction of the microbial community and may not represent the whole ARB community. This is another reason to advise against the use of E. coli to monitor AR. If a major aim of AR wastewater monitoring is to gain new insight into the dissemination of ARB&ARGs through the One-Health cycle (Hernando-Amado et al., Citation2019; McEwen & Collignon, Citation2018) E.coli will not permit an integrated surveillance. E. coli are not expected to occur in most human interfaces (drinking water, food and open spaces). Therefore, recommended targets are ARGs with common occurrence in wastewater and receiving environments and with clinical relevance. Because most environmental bacteria may be non-culturable, culture-independent methods are strongly recommended. These methods can be more informative, less laborious, and cheaper than culture-based approaches. The methods available so far to monitor AR fall within three major categories (). Because the assessment of treatment efficiency and of human’s transmission risks should be based on absolute quantifications, i.e., normalized per volume or mass of sample, culture-based or quantitative PCR (qPCR) methods are more adequate. However, all methods have advantages, and the best information, whenever feasible, should result from the combination of the three. The use of biomarkers that can be quantified based on routine methods is the best option for a regular and affordable monitoring scheme. The information available, summarized and discussed in a recent review (Keenum et al., Citation2022) shows the reliability and suitability of culture-independent methods, mainly of qPCR, to assess AR loads and provide comparative data (Table SI-1, ). A critical step to meet this goal is the definition of suitable biomarkers.
Table 2. Summary of most common methodological options for ARB/ARG monitoring in wastewater and related samples.
7.3. Goals and biomarkers: evidence-based decisions
The recommendation of regular AR monitoring in wastewater must be supported by the establishment of minimum requirements for discharges. One possibility of defining such requirements is the establishment of a threshold corresponding to the maximum ARG abundance that could be emitted by UWTPs. The current knowledge about occurrence, distribution, and risks associated with a specific ARG dose, as well as about the wastewater treatment processes required to guarantee such ARG dose, does not provide adequate background for such decision. Therefore, at the moment, a specific threshold cannot be recommended as the minimum requirement. An alternative approach to define an attainable goal is the proposal of minimum percentage of reduction. This type of goal has been in place for more than 30 years in the Urban Waste Water Directive (Council Directive, Citation1991, 91/271/EEC), with successful results, as have been recently published by the European Environmental Agency (https://www.eea.europa.eu/data-and-maps/indicators/urban-waste-water-treatment/urban-waste-water-treatment-assessment-5). In that directive, beside maximum admissible concentrations, also minimal percentage of reduction in relation to the load in the influent are recommended, for instance, for Biological Oxygen Demand (70–90%), Chemical Oxygen Demand (75%), Total Suspended Solids (90%), total nitrogen (70–80%) and total phosphorus (80%). This approach seems particularly adequate to establish goals of AR removal after wastewater treatment. Attainable percentage reduction values can be proposed based on the multiple studies available that show removal rates ranging from 1 to 3 log-units (per volume). Based on previous studies, the proposal of a minimum requirement of 95–99% (1.5–2.0 log-units), reduction of bacteria and indicator ARGs is a realistic and achievable goal that will contribute to reduce risks. Although if some regions may be prepared to implement more stringent requirements, an attainable objective may contribute to a measurable reduction of ARG emissions.
The proposal of suitable biomarkers must also rely on previous knowledge. Recent publications have discussed criteria and rankings to establish priority ARGs (Keenum et al., Citation2022; Zhang et al., Citation2021) in terms of human health risk and suitability to survey AR in surface water, recycled water and wastewater. The choice of the most suitable ARG candidates to monitor in water and wastewater is far from being a simple decision, being difficult to find common agreements. However, before listing ARGs, it may be helpful to establish some crucial criteria, specifically that the biomarker is:
Present in every sample suspected to contain contaminant ARGs with clinical relevance and associated to mobile genetic elements;
Abundant, permitting a reliable quantification, even after reduction due to wastewater treatment, dilution or other processes;
Stable in the environmental resistome, i.e., is part of the environmental ARG pool, and its distribution is not significantly increasing or reducing over time.
Some genetic determinants fulfill these criteria and may be proposed as suitable potential biomarkers (). Other are, for example, beta-lactam encoding resistance genes (e.g., blaTEM, blaOXA, blaSHV), tetracycline resistance genes (e.g., tetA, tet39, tetW, tetM or tetO), macrolide resistance (e.g., mefA), all frequently detected in UWTPs (Table SI-1; Keenum et al., Citation2022). Ideally a multi-biomarker system, including 5–10 ARGs or correlated MGEs and housekeeping genes, should be implemented. This process is feasible considering the current knowledge and technical developments.
Table 3. Examples of biomarkers that might be used to monitor AR removal in UWTPs, impacts on the environment, and to establish a first line of human-health risk assessment.
Monitoring methods can generate reproducible and comparable data in different laboratories (Rocha et al., Citation2019, Citation2020). The recent publication of guidelines for qPCR quality control, and the reporting of AR in wastewater and aquatic environment are excellent examples of the interest and feasibility of implementing interlaboratory monitoring schemes (Borchardt et al., Citation2021; Hassoun-Kheir et al. Citation2021; Keenum et al., Citation2022). Besides the increasing availability of real-time PCR and droplet digital PCR, the robustness of PCR primers design and protocols design, also the routinization and service providing for ARG analysis in environmental samples expanded in the last years (e.g., qiagen.com/∼/media/genetable/ba/antibioticresistancegenes; takarabio.com; resistomap.com). Commercial services offer high throughput methods that can be customized for the analysis of a selected number of genes in many samples. In general, ARG analyses will be increasingly feasible, customizable, and affordable. The body of knowledge about ARGs and related genetic elements in wastewater and receiving environments, as well as the technical availability to implement routine analysis capable of generating reproducible, objective and easy to interpret data, are strong arguments to put wastewater AR monitoring in place.
8. Final considerations
UWTPs are crucial to contain the spread of ARB&ARGs. AR monitoring framed within the set of quality parameters defined for treated wastewater will contribute to an improved knowledge of the best conditions to prevent its spread and will offer the operators the tools to optimize treatment conditions and infrastructure.
The definition of minimum quality requirements regarding the emission of ARGs by UWTPs would ideally reflect the risk posed to humans and the environment. However, the current scientific knowledge and technological wastewater treatment capabilities do not support yet the definition of science-evidence-based thresholds. Recent regulations (Table SI-2) show this same limitation. Hence, the definition of a minimum percentage reduction (>95–99%%) is an attainable goal that can be more stringent in countries that are already above this removal goal. Multiple ARG indicators measured in, at least the influent and final effluent of UWTPs, will support the determination of removal rates and impacts on the receiving environment (). Both can contribute to develop and implement corrective measures and through data sharing and metadata integration may support actions under the scope of One- and Global-Health (). Considering the current technological and scientific means available and the critical awareness worldwide of the threat posed by ARB&ARGs, the collaboration of all nations to implement monitoring and control systems is an urgent priority.
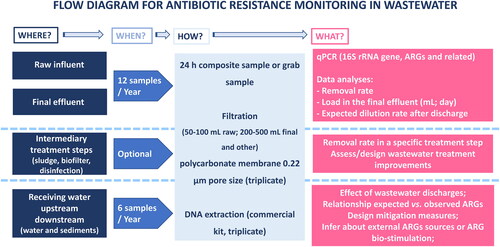
Figure 3. Proposed flow diagram for ARG monitoring in UWTPs to measure treatment efficiency, assess impacts, and to support the design of corrective interventions. Where, refers to preferential sampling sites; When, refers to periodicity of monitoring; How, summarizes the procedures; What, proposes what should be analyzed and the expected reporting outputs.
| Abbreviations | ||
| A2O | = | Anaerobic/anoxic/oxic |
| ARB | = | Antibiotic resistant bacteria |
| AR | = | Antibiotic resistance |
| ARGs | = | Antibiotic resistance genes |
| CW | = | Constructed wetlands |
| MIC | = | Minimal inhibitory concentrations |
| COD | = | Chemical Oxygen Demand |
| MGEs | = | Mobile genetic elements |
| p.e | = | Population equivalent |
| SI | = | Supporting information |
| UV | = | Ultra-violet |
| UWTPs | = | Urban wastewater treatment plants |
| UV | = | Ultra-violet radiation |
| MBR | = | Membrane bioreactors |
Supplemental Material
Download MS Word (692.9 KB)Additional information
Funding
References
- Akiyama, T., Asfahl, K. L., & Savin, M. C. (2010). Broad-host-range plasmids in treated wastewater effluent and receiving streams. Journal of Environmental Quality, 39(6), 2211–2215. https://doi.org/10.2134/jeq2010.0228
- Amos, G. C. A., Gozzard, E., Carter, C. E., Mead, A., Bowes, M. J., Hawkey, P. M., Zhang, L., Singer, A. C., Gaze, W. H., & Wellington, E. M. H. (2015). Validated predictive modelling of the environmental resistome. The ISME Journal, 9(6), 1467–1476. https://doi.org/10.1038/ismej.2014.237
- Amos, G. C. A., Hawkey, P. M., Gaze, W. H., & Wellington, E. M. (2014). Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. The Journal of Antimicrobial Chemotherapy, 69(7), 1785–1791. https://doi.org/10.1093/jac/dku079
- An, X.-L., Su, J.-Q., Li, B., Ouyang, W.-Y., Zhao, Y., Chen, Q.-L., Cui, L., Chen, H., Gillings, M. R., Zhang, T., & Zhu, Y.-G. (2018). Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR. Environment International, 117, 146–153. https://doi.org/10.1016/j.envint.2018.05.011
- Ashbolt, N., Pruden, A., Miller, J., Riquelme, M. V., & Maile-Moskowitz, A. (2018). Antimicrobal resistance: Fecal sanitation strategies for combatting a global public health threat. In: J. B. Rose & B. Jiménez-Cisneros, (Eds) Water and sanitation for the 21st century: Health and microbiological aspects of excreta and wastewater management (Global Water Pathogen Project). (A. Pruden, N. Ashbolt and J. Miller (eds), Part 3: Specific Excreted Pathogens: Environmental and Epidemiology Aspects - Section 2: Bacteria), Michigan State University, UNESCO. https://doi.org/10.14321/waterpathogens.29
- Becerra-Castro, C., Macedo, G., Silva, A. M. T., Manaia, C. M., & Nunes, O. C. (2016). Proteobacteria become predominant during regrowth after water disinfection. Science of the Total Environment, 573, 313–323. https://doi.org/10.1016/j.scitotenv.2016.08.054
- Bengtsson-palme, J., & Larsson, D. G. J. J. (2016). Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149. https://doi.org/10.1016/j.envint.2015.10.015
- Bengtsson-Palme, J., Hammarén, R., Pal, C., Östman, M., Björlenius, B., Flach, C.-F., Fick, J., Kristiansson, E., Tysklind, M., & Larsson, D. G. J. (2016). Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. The Science of the Total Environment, 572, 697–712. https://doi.org/10.1016/j.scitotenv.2016.06.228
- Bengtsson-Palme, J., Kristiansson, E., & Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42(1), fux053. https://doi.org/10.1093/femsre/fux053
- Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., Bürgmann, H., Sørum, H., Norström, M., Pons, M. N., Kreuzinger, N., Huovinen, P., Stefani, S., Schwartz, T., Kisand, V., Baquero, F., & Martinez, J. L. (2015). Tackling antibiotic resistance: The environmental framework. Nature Reviews. Microbiology, 13(5), 310–317. https://doi.org/10.1038/nrmicro3439
- Borchardt, M. A., Boehm, A. B., Salit, M., Spencer, S. K., Wigginton, K. R., & Noble, R. T. (2021). The Environmental microbiology minimum information (EMMI) Guidelines: qPCR and dPCR quality and reporting for environmental microbiology. Environmental Science & Technology, 55(15), 10210–10223. https://doi.org/10.1021/acs.est.1c01767
- Brown, P. C., Borowska, E., Schwartz, T., & Horn, H. (2019). Impact of the particulate matter from wastewater discharge on the abundance of antibiotic resistance genes and facultative pathogenic bacteria in downstream river sediments. Science of the Total Environment, 649, 1171–1178. https://doi.org/10.1016/j.scitotenv.2018.08.394
- Cacace, D., Fatta-Kassinos, D., Manaia, C. M., Cytryn, E., Kreuzinger, N., Rizzo, L., Karaolia, P., Schwartz, T., Alexander, J., Merlin, C., Garelick, H., Schmitt, H., de Vries, D., Schwermer, C. U., Meric, S., Ozkal, C. B., Pons, M. N., Kneis, D., & Berendonk, T. U. (2019). Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Research, 162, 320–330. https://doi.org/10.1016/j.watres.2019.06.039
- Chen, H., & Zhang, M. (2013). Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environmental Science & Technology, 47(15), 8157–8163. https://doi.org/10.1021/es401091y
- Chu, B. T. T., Petrovich, M. L., Chaudhary, A., Wright, D., Murphy, B., Wells, G., & Poretsky, R. (2018). Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. Applied and Environmental Microbiology, 84(5), e02168–17. https://doi.org/10.1128/AEM.02168-17
- Council Directive. (1991). Council Directive of 21 concerning urban waste water treatment (91/271/EEC), May.
- Czekalski, N., Sigdel, R., Birtel, J., Matthews, B., & Bürgmann, H. (2015). Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environment International, 81, 45–55. https://doi.org/10.1016/j.envint.2015.04.005
- Dai, D., Brown, C., Bürgmann, H., Larsson, D. G. J., Nambi, I., Zhang, T., Flach, C. F., Pruden, A., & Vikesland, P. J. (2022). Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements from sewage to activated sludge. Microbiome, 10(1), 20. https://doi.org/10.1186/s40168-021-01216-5
- Davies, J., & Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews: MMBR, 74(3), 417–433. https://doi.org/10.1128/MMBR.00016-10
- Devarajan, N., Laffite, A., Graham, N. D., Meijer, M., Prabakar, K., Mubedi, J. I., Elongo, V., Mpiana, P. T., Ibelings, B. W., Wildi, W., & Poté, J. (2015). Accumulation of clinically relevant antibiotic-resistance genes, bacterial load, and metals in freshwater lake sediments in Central Europe. Environmental Science & Technology, 49(11), 6528–6537. https://doi.org/10.1021/acs.est.5b01031
- Di Cesare, A., Fontaneto, D., Doppelbauer, J., & Corno, G. (2016). Fitness and recovery of bacterial communities and antibiotic resistance genes in urban wastewaters exposed to classical disinfection treatments. Environmental Science & Technology, 50(18), 10153–10161. https://doi.org/10.1021/acs.est.6b02268
- Ding, H., Qiao, M., Zhong, J., Zhu, Y., Guo, C., Zhang, Q., Yang, P., Han, L., Zhang, W., Wu, Y., Liu, J., Zhang, L., & Sun, J. (2020). Characterization of antibiotic resistance genes and bacterial community in selected municipal and industrial sewage treatment plants beside Poyang Lake. Water Research, 174, 115603. https://doi.org/10.1016/j.watres.2020.115603
- Du, J., Geng, J., Ren, H., Ding, L., Xu, K., & Zhang, Y. (2015). Variation of antibiotic resistance genes in municipal wastewater treatment plant with A(2)O-MBR system. Environmental Science and Pollution Research International, 22(5), 3715–3726. https://doi.org/10.1007/s11356-014-3552-x
- Fan, Y., Guo, Z., Wang, J., Zhang, B., Shen, Y., & Gao, X. (2022). Online learning-empowered smart management for A2O process in sewage treatment processes. Environmental Research, 210, 113015. https://doi.org/10.1016/j.envres.2022.113015
- Ferreira da Silva, M., Vaz-Moreira, I., Gonzalez-Pajuelo, M., Nunes, O. C., & Manaia, C. M. (2007). Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiology Ecology, 60(1), 166–176. https://doi.org/10.1111/j.1574-6941.2006.00268.x
- Fiorentino, A., Di Cesare, A., Eckert, E. M., Rizzo, L., Fontaneto, D., Yang, Y., & Corno, G. (2019). Impact of industrial wastewater on the dynamics of antibiotic resistance genes in a full-scale urban wastewater treatment plant. The Science of the Total Environment, 646, 1204–1210. https://doi.org/10.1016/j.scitotenv.2018.07.370
- Freeman, C. N., Scriver, L., Neudorf, K. D., Hansen, L. T., Jamieson, R. C., & Yost, C. K. (2018). Antimicrobial resistance gene surveillance in the receiving waters of an upgraded wastewater treatment plant. FACETS, 3(1), 128–138. https://doi.org/10.1139/facets-2017-0085
- Garner, E., Wallace, J. S., Argoty, G. A., Wilkinson, C., Fahrenfeld, N., Heath, L. S., Zhang, L., Arabi, M., Aga, D. S., & Pruden, A. (2016). Metagenomic profiling of historic Colorado Front Range flood impact on distribution of riverine antibiotic resistance genes. Scientific Reports, 6, 38432. https://doi.org/10.1038/srep38432
- Gaze, W. H., Zhang, L., Abdouslam, N. A., Hawkey, P. M., Calvo-Bado, L., Royle, J., Brown, H., Davis, S., Kay, P., Boxall, A. B., & Wellington, E. M. (2011). Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. The ISME Journal, 5(8), 1253–1261. https://doi.org/10.1038/ismej.2011.15
- Gibson, B., Wilson, D. J., Feil, E., & Eyre-Walker, A. (2018). The distribution of bacterial doubling times in the wild. Proceedings of the Royal Society B: Biological Sciences, 285(1880), 20180789. https://doi.org/10.1098/rspb.2018.0789
- Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., & Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. The ISME Journal, 9(6), 1269–1279. https://doi.org/10.1038/ismej.2014.226
- Guerin, E., Cambray, G., Sanchez-Alberola, N., Campoy, S., Erill, I., Da Re, S., Gonzalez-Zorn, B., Barbé, J., Ploy, M. C., & Mazel, D. (2009). The SOS response controls integron recombination. Science (New York, N.Y.), 324(5930), 1034. https://doi.org/10.1126/science.1172914
- Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., & Andersson, D. I. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathogens, 7(7), e1002158. https://doi.org/10.1371/journal.ppat.1002158
- Harnisz, M., Kiedrzyńska, E., Kiedrzyński, M., Korzeniewska, E., Czatzkowska, M., Koniuszewska, I., Jóźwik, A., Szklarek, S., Niestępski, S., & Zalewski, M. (2020). The impact of WWTP size and sampling season on the prevalence of antibiotic resistance genes in wastewater and the river system. Science of the Total Environment, 741, 140466. https://doi.org/10.1016/j.scitotenv.2020.140466
- Hassoun-Kheir, N., Stabholz, Y., Kreft, J. U., de la Cruz, R., Dechesne, A., Smets, B. F., Romalde, J. L., Lema, A., Balboa, S., García-Riestra, C., Torres-Sangiao, E., Neuberger, A., Graham, D., Quintela-Baluja, M., Stekel, D. J., Graham, J., Pruden, A., Nesme, J., Sørensen, S. J., Hough, R., & Paul, M. (2021). EMBRACE-WATERS statement: Recommendations for reporting of studies on antimicrobial resistance in wastewater and related aquatic environments. One Health (Amsterdam, Netherlands), 13, 100339. https://doi.org/10.1016/j.onehlt.2021.100339
- Hernando-Amado, S., Coque, T. M., Baquero, F., & Martínez, J. L. (2019). Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nature Microbiology, 4(9), 1432–1442. https://doi.org/10.1038/s41564-019-0503-9
- Hiller, C. X., Hübner, U., Fajnorova, S., Schwartz, T., & Drewes, J. E. (2019). Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. The Science of the Total Environment, 685, 596–608. https://doi.org/10.1016/j.scitotenv.2019.05.315
- Honda, R., Tachi, C., Noguchi, M., Yamamoto-Ikemoto, R., & Watanabe, T. (2020). Fate and seasonal change of Escherichia coli resistant to different antibiotic classes at each stage of conventional activated sludge process. Journal of Water and Health, 18(6), 879–889. https://doi.org/10.2166/wh.2020.013
- Hubeny, J., Harnisz, M., Korzeniewska, E., Buta, M., Zieliński, W., Rolbiecki, D., Giebułtowicz, J., Nałęcz-Jawecki, G., & Płaza, G. (2021). Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PLoS ONE, 16(6), e0252691. https://doi.org/10.1371/journal.pone.0252691
- Huijbers, P. M. C., Blaak, H., de Jong, M. C. M., Graat, E. A. M., Vandenbroucke-Grauls, C. M. J. E., & de Roda Husman, A. M. (2015). Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environmental Science & Technology, 49(20), 11993–12004. https://doi.org/10.1021/acs.est.5b02566
- Hutinel, M., Huijbers, P. M. C., Fick, J., Åhrén, C., Larsson, D. G. J., & Flach, C. F. (2019). Population-level surveillance of antibiotic resistance in Escherichia coli through sewage analysis. Eurosurveillance, 24(37), 1800497. https://doi.org/10.2807/1560-7917.ES.2019.24.37.1800497
- Iakovides, I. C., Manoli, K., Karaolia, P., Michael-Kordatou, I., Manaia, C. M., & Fatta-Kassinos, D. (2021). Reduction of antibiotic resistance determinants in urban wastewater by ozone: Emphasis on the impact of wastewater matrix towards the inactivation kinetics, toxicity and bacterial regrowth. Journal of Hazardous Materials, 420, 126527. https://doi.org/10.1016/j.jhazmat.2021.126527
- Jia, S., Li, T., & Zhang, X. X. (2021). Integrated metagenomic and metatranscriptomic analyses of ultraviolet disinfection effects on antibiotic resistance genes and bacterial communities during wastewater treatment. Ecotoxicology, 30(8), 1610–1619. https://doi.org/10.1007/s10646-020-02313-1
- Kalinowska, A., Jankowska, K., Fudala-Ksiazek, S., Pierpaoli, M., & Luczkiewicz, A. (2021). The microbial community, its biochemical potential, and the antimicrobial resistance of Enterococcus spp. in Arctic lakes under natural and anthropogenic impact (West Spitsbergen). The Science of the Total Environment, 763, 142998. https://doi.org/10.1016/j.scitotenv.2020.142998
- Karkman, A., Pärnänen, K., & Larsson, D. G. J. (2019). Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nature Communications, 10(1), 1–8. https://doi.org/10.1038/s41467-018-07992-3
- Keenum, I., Liguori, K., Calarco, J., Davis, B. C., Milligan, E., Harwood, V. J., & Pruden, A. (2022). A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Critical Reviews in Environmental Science and Technology, 1–25. https://doi.org/10.1080/10643389.2021.2024739
- Keller, V. D., Williams, R. J., Lofthouse, C., & Johnson, A. C. (2014). Worldwide estimation of river concentrations of any chemical originating from sewage-treatment plants using dilution factors. Environmental Toxicology and Chemistry, 33(2), 447–452. https://doi.org/10.1002/etc.2441
- Klümper, U., Recker, M., Zhang, L., Yin, X., Zhang, T., Buckling, A., & Gaze, W. H. (2019). Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. The ISME Journal, 13(12), 2927–2937. https://doi.org/10.1038/s41396-019-0483-z
- Korajkic, A., McMinn, B. R., & Harwood, V. J. (2018). Relationships between microbial indicators and pathogens in recreational water settings. International Journal of Environmental Research and Public Health, 15(12), 2842. https://doi.org/10.3390/ijerph15122842
- Krzeminski, P., Tomei, M. C., Karaolia, P., Langenhoff, A., Almeida, C. M. R., Felis, E., Gritten, F., Andersen, H. R., Fernandes, T., Manaia, C. M., Rizzo, L., & Fatta-Kassinos, D. (2019). Performance of secondary wastewater treatment methods for the removal of contaminants of emerging concern implicated in crop uptake and antibiotic resistance spread: A review. The Science of the Total Environment, 648, 1052–1081. https://doi.org/10.1016/j.scitotenv.2018.08.130
- Laht, M., Karkman, A., Voolaid, V., Ritz, C., Tenson, T., Virta, M., & Kisand, V. (2014). Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS ONE, 9(8), e103705. https://doi.org/10.1371/journal.pone.0103705
- LaPara, T. M., Burch, T. R., McNamara, P. J., Tan, D. T., Yan, M., & Eichmiller, J. J. (2011). Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environmental Science & Technology, 45(22), 9543–9549. https://doi.org/10.1021/es202775r
- Le, T. H., Ng, C., Tran, N. H., Chen, H., & Gin, K. Y. H. (2018). Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Research, 145, 498–508. https://doi.org/10.1016/j.watres.2018.08.060
- Lee, J., Jeon, J. H., Shin, J., Jang, H. M., Kim, S., Song, M. S., & Kim, Y. M. (2017). Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. The Science of the Total Environment, 605–606, 906–914. https://doi.org/10.1016/j.scitotenv.2017.06.250
- Lehmann, K., Bell, T., Bowes, M. J., Amos, G. C. A., Gaze, W. H., Wellington, E. M. H., & Singer, A. C. (2016). Trace levels of sewage effluent are sufficient to increase class 1 integron prevalence in freshwater biofilms without changing the core community. Water Research, 106, 163–170. https://doi.org/10.1016/j.watres.2016.09.035
- Lekunberri, I., Balcázar, J. L., & Borrego, C. M. (2018). Metagenomic exploration reveals a marked change in the river resistome and mobilome after treated wastewater discharges. Environmental Pollution (Barking, Essex : 1987), 234, 538–542. https://doi.org/10.1016/j.envpol.2017.12.001
- Lekunberri, I., Villagrasa, M., Balcázar, J. L., & Borrego, C. M. (2017). Contribution of bacteriophage and plasmid DNA to the mobilization of antibiotic resistance genes in a river receiving treated wastewater discharges. Science of the Total Environment, 601-602, 206–209. https://doi.org/10.1016/j.scitotenv.2017.05.174
- Leonard, A. F. C., Yin, X. L., Zhang, T., Hui, M., & Gaze, W. H. (2018). A coliform-targeted metagenomic method facilitating human exposure estimates to Escherichia coli-borne antibiotic resistance genes. FEMS Microbiology Ecology, 94(3). https://doi.org/10.1093/femsec/fiy024
- Leonard, A. F. C., Zhang, L., Balfour, A. J., Garside, R., Hawkey, P. M., Murray, A. K., Ukoumunne O. C., & Gaze W. H. (2018). Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: Environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environment International, 114, 326–333. http://dx.doi.org/10.1016/j.envint.2017.11.003
- Lin, X., Ruan, J., Huang, L., Zhao, J., & Xu, Y. (2021). Comparison of the elimination effectiveness of tetracycline and AmpC β-lactamase resistance genes in a municipal wastewater treatment plant using four parallel processes. Ecotoxicology, 30(8), 1586–1597. https://doi.org/10.1007/s10646-020-02306-0
- Lin, Z.-J., Zhou, Z.-C., Zhu, L., Meng, L.-X., Shuai, X.-Y., Sun, Y.-J., & Chen, H. (2021). Behavior of antibiotic resistance genes in a wastewater treatment plant with different upgrading processes. Science of the Total Environment, 771, 144814. https://doi.org/10.1016/j.scitotenv.2020.144814
- Lira, F., Vaz-Moreira, I., Tamames, J., Manaia, C. M., & Martínez, J. L. (2020). Metagenomic analysis of an urban resistome before and after wastewater treatment. Scientific Reports, 10(1), 8174. https://doi.org/10.1038/s41598-020-65031-y
- Liu, Y., Liu, W., Yang, X., Wang, J., Lin, H., & Yang, Y. (2021). Microplastics are a hotspot for antibiotic resistance genes: Progress and perspective. The Science of the Total Environment, 773(1), 145643. https://doi.org/10.1016/j.scitotenv.2021.145643
- Liu, S.-S., Qu, H.-M., Yang, D., Hu, H., Liu, W.-L., Qiu, Z.-G., Hou, A.-M., Guo, J., Li, J.-W., Shen, Z.-Q., & Jin, M. (2018). Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Research, 136, 131–136. https://doi.org/10.1016/j.watres.2018.02.036
- Liu, Y., Tong, Z., Shi, J., Jia, Y., Yang, K., & Wang, Z. (2020). Correlation between exogenous compounds and the horizontal transfer of plasmid-borne antibiotic resistance genes. Microorganisms, 8(8), 1211. https://doi.org/10.3390/microorganisms8081211
- Li, L. G., Xia, Y., & Zhang, T. (2017). Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. The ISME Journal, 11(3), 651–662. https://doi.org/10.1038/ismej.2016.155
- Li, L. G., Yin, X., & Zhang, T. (2018). Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome, 6(1), 93. https://doi.org/10.1186/s40168-018-0480-x
- Ma, L., Li, A. D., Yin, X. L., & Zhang, T. (2017). The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environmental Science & Technology, 51(10), 5721–5728. https://doi.org/10.1021/acs.est.6b05887
- Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: Non-direct proportionality between abundance and risk. Trends in Microbiology, 25(3), 173–181. https://doi.org/10.1016/j.tim.2016.11.014
- Marano, R. B., Fernandes, T., Manaia, C. M., Nunes, O., Morrison, D., Berendonk, T. U., Kreuzinger, N., Tenson, T., Corno, G., Fatta-Kassinos, D., Merlin, C., Topp, E., Jurkevitch, E., Henn, L., Scott, A., Heß, S., Slipko, K., Laht, M., Kisand, V., … Cytryn, E. (2020). A global multinational survey of cefotaxime-resistant coliforms in urban wastewater treatment plants. Environment International, 144, 106035. https://doi.org/10.1016/j.envint.2020.106035
- Marti, E., Jofre, J., & Balcazar, J. L. (2013). Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE, 8(10), e78906. https://doi.org/10.1371/journal.pone.0078906
- McConnell, M. M., Hansen, L. T., Neudorf, K. D., Hayward, J. L., Jamieson, R. C., Yost, C. K., & Tong, A. (2018a). Sources of antibiotic resistance genes in a rural river system. Journal of Environmental Quality, 47(5), 997–1005. https://doi.org/10.2134/jeq2017.12.0477
- McConnell, M. M., Truelstrup Hansen, L., Jamieson, R. C., Neudorf, K. D., Yost, C. K., & Tong, A. (2018b). Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Science of the Total Environment, 643, 292–300. https://doi.org/10.1016/j.scitotenv.2018.06.212
- McEwen, S. A., & Collignon, P. J. (2018). Antimicrobial resistance: A one health perspective. Microbiology Spectrum, 6(2). https://doi.org/10.1128/microbiolspec.ARBA-0009-2017
- McGough, S. F., MacFadden, D. R., Hattab, M. W., Mølbak, K., & Santillana, M. (2020). Rates of increase of antibiotic resistance and ambient temperature in Europe: A cross-national analysis of 28 countries between 2000 and 2016. Eurosurveillance, 25(45), 1–12. https://doi.org/10.2807/1560-7917.ES.2020.25.45.1900414
- Miłobedzka, A., Ferreira, C., Vaz-Moreira, I., Calderón-Franco, D., Gorecki, A., Purkrtova, S., Bartacek, J., Dziewit, L., Singleton, C. M., Nielsen, P. H., Weissbrodt, D. G., & Manaia, C. M. (2022). Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the one-health cycle. Journal of Hazardous Materials, 424(Pt C), 127407. https://doi.org/10.1016/j.jhazmat.2021.127407
- Moreira, N. F. F., Narciso-da-Rocha, C., Polo-López, M. I., Pastrana-Martínez, L. M., Faria, J. L., Manaia, C. M., Fernández-Ibáñez, P., Nunes, O. C., & Silva, A. M. T. (2018). Solar treatment (H2O2, TiO2-P25 and GO-TiO2 photocatalysis, photo-Fenton) of organic micropollutants, human pathogen indicators, antibiotic resistant bacteria and related genes in urban wastewater. Water Research, 135, 195–206. https://doi.org/10.1016/j.watres.2018.01.064
- Mughini-Gras, L., Dorado-García, A., van Duijkeren, E., van den Bunt, G., Dierikx, C. M., Bonten, M. J. M., Bootsma, M. C. J., Schmitt, H., Hald, T., Evers, E. G., de Koeijer, A., van Pelt, W., Franz, E., Mevius, D. J., & Heederik, D. J. J. (2019). Heederik DJJ; ESBL Attribution Consortium. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. The Lancet. Planetary Health, 3(8), e357–e369. Erratum in: Lancet Planet Health. 2019 Oct;3(10):e408. https://doi.org/10.1016/S2542-5196(19)30130-5
- Munck, C., Albertsen, M., Telke, A., Ellabaan, M., Nielsen, P. H., & Sommer, M. O. A. (2015). Limited dissemination of the wastewater treatment plant core resistome. Nature Communications, 6(1), 8452. https://doi.org/10.1038/ncomms9452
- Murray, A. K., Stanton, I., Gaze, W. H., & Snape, J. (2021). Dawning of a new ERA: Environmental risk assessment of antibiotics and their potential to select for antimicrobial resistance. Water Research, 200, 117233. https://doi.org/10.1016/j.watres.2021.117233
- Narciso-da-Rocha, C., Rocha, J., Vaz-Moreira, I., Lira, F., Tamames, J., Henriques, I., Martinez, J. L., & Manaia, C. M. (2018). Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environment International, 118(June), 179–188. https://doi.org/10.1016/j.envint.2018.05.040
- Narciso-da-Rocha, C., Varela, A. R., Schwartz, T., Nunes, O. C., & Manaia, C. M. (2014). blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital-urban wastewater treatment plant system. Journal of Global Antimicrobial Resistance, 2(4), 309–315. https://doi.org/10.1016/j.jgar.2014.10.001
- Ng, C., Tan, B., Jiang, X. T., Gu, X., Chen, H., Schmitz, B. W., Haller, L., Charles, F. R., Zhang, T., & Gin, K. (2019). Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in singapore containing membrane bioreactors. Frontiers in Microbiology, 10, 172. https://doi.org/10.3389/fmicb.2019.00172
- Novo, A., André, S., Viana, P., Nunes, O. C., & Manaia, C. M. (2013). Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Research, 47(5), 1875–1887. https://doi.org/10.1016/j.watres.2013.01.010
- Novo, A., & Manaia, C. M. (2010). Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Applied Microbiology and Biotechnology, 87(3), 1157–1166. https://doi.org/10.1007/s00253-010-2583-6
- Oliveira, M., Nunes, M., Barreto Crespo, M. T., & Silva, A. F. (2020). The environmental contribution to the dissemination of carbapenem and (fluoro)quinolone resistance genes by discharged and reused wastewater effluents: The role of cellular and extracellular DNA. Water Research, 182, 116011. https://doi.org/10.1016/j.watres.2020.116011
- Pallares-Vega, R., Blaak, H., van der Plaats, R., de Roda Husman, A. M., Hernandez Leal, L., van Loosdrecht, M. C., Weissbrodt, D. G., & Schmitt, H. (2019). Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: A cross-sectional study. Water Research, 161, 319–328. https://doi.org/10.1016/j.watres.2019.05.100
- Pallares-Vega, R., Hernandez Leal, L., Fletcher, B. N., Vias-Torres, E., van Loosdrecht, M. C., Weissbrodt, D. G., & Schmitt, H. (2021). Annual dynamics of antimicrobials and resistance determinants in flocculent and aerobic granular sludge treatment systems. Water Research, 190, 116752. https://doi.org/10.1016/j.watres.2020.116752
- Park, G. W., Ng, T. F. F., Freeland, A. L., Marconi, V. C., Boom, J. A., Staat, M. A., Montmayeur, A. M., Browne, H., Narayanan, J., Payne, D. C., Cardemil, C. V., Treffiletti, A., & Vinjé, J. (2020). CrAssphage as a novel tool to detect human fecal contamination on environmental surfaces and hands. Emerging Infectious Diseases, 26(8), 1731–1739. https://doi.org/10.3201/eid2608.200346
- Pärnänen, K. M. M., Narciso-da-Rocha, C., Kneis, D., Berendonk, T. U., Cacace, D., & Do, T. T. (2019). Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Science Advances, 5(3), eaau9124. http://advances.sciencemag.org/content/5/3/eaau9124
- Paul, D., Chakraborty, R., & Mandal, S. M. (2019). Biocides and health-care agents are more than just antibiotics: Inducing cross to co-resistance in microbes. Ecotoxicology and Environmental Safety, 174(March), 601–610. https://doi.org/10.1016/j.ecoenv.2019.02.083
- Pei, M., Zhang, B., He, Y., Su, J., Gin, K., Lev, O., Shen, G., & Hu, S. (2019). State of the art of tertiary treatment technologies for controlling antibiotic resistance in wastewater treatment plants. Environment International, 131(March), 105026. https://doi.org/10.1016/j.envint.2019.105026
- Proia, L., Anzil, A., Borrego, C., Farrè, M., Llorca, M., Sanchis, J., Bogaerts, P., Balcázar, J. L., & Servais, P. (2018a). Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. Journal of Hazardous Materials, 358(June), 33–43. https://doi.org/10.1016/j.jhazmat.2018.06.058
- Proia, L., Anzil, A., Subirats, J., Borrego, C., Farrè, M., Llorca, M., Balcázar, J. L., & Servais, P. (2018b). Antibiotic resistance along an urban river impacted by treated wastewaters. The Science of the Total Environment, 628-629, 453–466. https://doi.org/10.1016/j.scitotenv.2018.02.083
- Qiu, Z., Yu, Y., Chen, Z., Jin, M., Yang, D., Zhao, Z., Wang, J., Shen, Z., Wang, X., Qian, D., Huang, A., Zhang, B., & Li, J. W. (2012). Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proceedings of the National Academy of Sciences of the United States of America, 109(13), 4944–4949. https://doi.org/10.1073/pnas.1107254109
- Quintela-Baluja, M., Abouelnaga, M., Romalde, J., Su, J.-Q., Yu, Y., Gomez-Lopez, M., Smets, B., Zhu, Y.-G., & Graham, D. W. (2019). Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters. Water Research, 162, 347–357. https://doi.org/10.1016/j.watres.2019.06.075
- Raza, S., Jo, H., Kim, J., Shin, H., Hur, H. G., & Unno, T. (2021). Metagenomic exploration of antibiotic resistome in treated wastewater effluents and their receiving water. The Science of the Total Environment, 765, 142755. https://doi.org/10.1016/j.scitotenv.2020.142755
- Ribeirinho-Soares, S., Moreira, N. F. F., Graça, C., Pereira, M. F. R., Silva, A. M. T., & Nunes, O. C. (2021). Overgrowth control of potentially hazardous bacteria during storage of ozone treated wastewater through natural competition. Water Research, 209, 117932. https://doi.org/10.1016/j.watres.2021.117932
- Rilstone, V., Vignale, L., Craddock, J., Cushing, A., Filion, Y., & Champagne, P. (2021). The role of antibiotics and heavy metals on the development, promotion, and dissemination of antimicrobial resistance in drinking water biofilms. Chemosphere, 282(June), 131048. https://doi.org/10.1016/j.chemosphere.2021.131048
- Rizzo, L., Gernjak, W., Krzeminski, P., Malato, S., McArdell, C. S., Perez, J. A. S., Schaar, H., & Fatta-Kassinos, D. (2020). Best available technologies and treatment trains to address current challenges in urban wastewater reuse for irrigation of crops in EU countries. The Science of the Total Environment, 710, 136312. https://doi.org/10.1016/j.scitotenv.2019.136312
- Rocha, J., Cacace, D., Kampouris, I., Guilloteau, H., Jäger, T., & Marano, R. B. M. (2020). Inter-laboratory calibration of quantitative analyses of antibiotic resistance genes. Journal of Environmental Chemical Engineering, 8(1), 102214. https://doi.org/10.1016/j.jece.2018.02.022
- Rocha, J., Fernandes, T., Riquelme, M. V., Zhu, N., Pruden, A., & Manaia, C. M. (2019). Comparison of culture- and quantitative PCR-based indicators of antibiotic resistance in wastewater, recycled water, and tap water. International Journal of Environmental Research and Public Health, 16(21), 4217. https://doi.org/10.3390/ijerph16214217
- Rodriguez-Mozaz, S., Chamorro, S., Marti, E., Huerta, B., Gros, M., Sànchez-Melsió, A., Borrego, C. M., Barceló, D., & Balcázar, J. L. (2015). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Research, 69, 234–242. https://doi.org/10.1016/j.watres.2014.11.021
- Rodriguez-Mozaz, S., Vaz-Moreira, I., Varela Della Giustina, S., Llorca, M., Barceló, D., Schubert, S., Berendonk, T. U., Michael-Kordatou, I., Fatta-Kassinos, D., Martinez, J. L., Elpers, C., Henriques, I., Jaeger, T., Schwartz, T., Paulshus, E., O'Sullivan, K., Pärnänen, K. M. M., Virta, M., Do, T. T., Walsh, F., & Manaia, C. M. (2020). Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environment International, 140, 105733. https://doi.org/10.1016/j.envint.2020.105733
- Saxena, P., Hiwrale, I., Das, S., Shukla, V., Tyagi, L., Pal, S., Dafale, N., & Dhodapkar, R. (2021). Profiling of emerging contaminants and antibiotic resistance in sewage treatment plants: An Indian perspective. Journal of Hazardous Materials, 408, 124877. https://doi.org/10.1016/j.jhazmat.2020.124877
- Sousa, J. M., Macedo, G., Pedrosa, M., Becerra-Castro, C., Castro-Silva, S., Pereira, M. F. R., Silva, A. M. T., Nunes, O. C., & Manaia, C. M. (2017). Ozonation and UV254nm radiation for the removal of microorganisms and antibiotic resistance genes from urban wastewater. Journal of Hazardous Materials, 323(Pt A), 434–441. https://doi.org/10.1016/j.jhazmat.2016.03.096
- Storteboom, H., Arabi, M., Davis, J. G., Crimi, B., & Pruden, A. (2010). Tracking antibiotic resistance genes in the South Platte River basin using molecular signatures of urban, agricultural, and pristine sources. Environmental Science & Technology, 44(19), 7397–7404. https://doi.org/10.1021/es101657s
- Tello, A., Austin, B., & Telfer, T. C. (2012). Selective pressure of antibiotic pollution on bacteria of importance to public health. Environmental Health Perspectives, 120(8), 1100–1106. https://doi.org/10.1289/ehp.1104650
- Thakali, O., Brooks, J. P., Shahin, S., Sherchan, S. P., & Haramoto, E. (2020). Removal of antibiotic resistance genes at two conventional wastewater treatment plants of Louisiana, USA. Water (Switzerland), 12(6), 1729. https://doi.org/10.3390/w12061729
- Thornton, C. N., Tanner, W. D., VanDerslice, J. A., & Brazelton, W. J. (2020). Localized effect of treated wastewater effluent on the resistome of an urban watershed. GigaScience, 9(11), giaa125. https://doi.org/10.1093/gigascience/giaa125
- Tong, C., Hu, H., Chen, G., Li, Z., Li, A., & Zhang, J. (2021). Disinfectant resistance in bacteria: Mechanisms, spread, and resolution strategies. Environmental Research, 195, 110897. https://doi.org/10.1016/j.envres.2021.110897
- Tong, J., Tang, A., Wang, H., Liu, X., Huang, Z., Wang, Z., Zhang, J., Wei, Y., Su, Y., & Zhang, Y. (2019). Microbial community evolution and fate of antibiotic resistance genes along six different full-scale municipal wastewater treatment processes. Bioresource Technology, 272, 489–500. https://doi.org/10.1016/j.biortech.2018.10.079
- UN-Water. (2020). Summary Progress Update 2021 – SDG 6 – water and sanitation for all. Version: 1 March 2021.
- Varela, A. R., André, S., Nunes, O. C., & Manaia, C. M. (2014). Insights into the relationship between antimicrobial residues and bacterial populations in a hospital-urban wastewater treatment plant system . Water Research, 54, 327–336. https://doi.org/10.1016/j.watres.2014.02.003
- Vaz-Moreira, I., Nunes, O. C., & Manaia, C. M. (2014). Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiology Reviews, 38(4), 761–778. https://doi.org/10.1111/1574-6976.12062
- Vilanova, X., Manero, A., Cerdà-Cuéllar, M., & Blanch, A. R. (2004). The composition and persistence of faecal coliforms and enterococcal populations in sewage treatment plants. Journal of Applied Microbiology, 96(2), 279–288. https://doi.org/10.1046/j.1365-2672.2003.02149.x
- Vos, M., Sibleyras, L., Lo, L. K., Hesse, E., Gaze, W., & Klümper, U. (2020). Zinc can counteract selection for ciprofloxacin resistance. FEMS Microbiology Letters, 367(3), fnaa038. https://doi.org/10.1093/femsle/fnaa038
- Wang, Y., Lu, J., Mao, L., Li, J., Yuan, Z., Bond, P. L., & Guo, J. (2019). Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. The ISME Journal, 13(2), 509–522. https://doi.org/10.1038/s41396-018-0275-x
- Wang, M., Shen, W., Yan, L., Wang, X. H., & Xu, H. (2017). Stepwise impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environmental Pollution, 231(Pt 2), 1578–1585. https://doi.org/10.1016/j.envpol.2017.09.055
- Webber, M. A., & Piddock, L. J. (2003). The importance of efflux pumps in bacterial antibiotic resistance. The Journal of Antimicrobial Chemotherapy, 51(1), 9–11. https://doi.org/10.1093/jac/dkg050
- Wen, Q., Yang, L., Duan, R., & Chen, Z. (2016). Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China. Environmental Pollution (Barking, Essex: 1987), 212, 34–40. https://doi.org/10.1016/j.envpol.2016.01.043
- World Health Organization. (2019). Newsroom/Spotlight/Ten threats to global health in 2019. https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019.
- World Health Organization. (2021). Integrated global surveillance on ESBL-producing E. coli using a “One Health” approach: Implementation and opportunities. World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
- Yang, Y., Liu, G., Ye, C., & Liu, W. (2019b). Bacterial community and climate change implication affected the diversity and abundance of antibiotic resistance genes in wetlands on the Qinghai-Tibetan Plateau. Journal of Hazardous Materials, 361(1), 283–293. https://doi.org/10.1016/j.jhazmat.2018.09.002
- Yang, L., Wen, Q., Chen, Z., Duan, R., & Yang, P. (2019a). Impacts of advanced treatment processes on elimination of antibiotic resistance genes in a municipal wastewater treatment plant. Frontiers of Environmental Science & Engineering, 13, 32. https://doi.org/10.1007/s11783-019-1116-5
- Yi, T., Kim, T. G., & Cho, K. S. (2015). Fate and behavior of extended-spectrum β-lactamase-producing genes in municipal sewage treatment plants. Journal of Environmental Science and Health. Part A, Toxic/Hazardous Substances & Environmental Engineering, 50(11), 1160–1168. https://doi.org/10.1080/10934529.2015.1047673
- Yu, K., Li, P., He, Y., Zhang, B., Chen, Y., & Yang, J. (2020). Unveiling dynamics of size-dependent antibiotic resistome associated with microbial communities in full-scale wastewater treatment plants. Water Research, 187, 116450. https://doi.org/10.1016/j.watres.2020.116450
- Zhang, A. N., Gaston, J. M., Dai, C. L., Zhao, S., Poyet, M., Groussin, M., Yin, X., Li, L. G., van Loosdrecht, M. C. M., Topp, E., Gillings, M. R., Hanage, W. P., Tiedje, J. M., Moniz, K., Alm, E. J., & Zhang, T. (2021). An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nature Communications, 12(1), 4765. https://doi.org/10.1038/s41467-021-25096-3
- Zhang, Y., Gu, A. Z., Cen, T., Li, X., He, M., Li, D., & Chen, J. (2018). Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environmental Pollution (Barking, Essex : 1987), 237, 74–82. https://doi.org/10.1016/j.envpol.2018.01.032
- Zhang, S., Lv, L., Zhang, Y., Zhang, H., Yu, X., & Zhang, S. (2013). Occurrence and variations of five classes of antibiotic resistance genes along the Jiulong River in southeast China. Journal of Environmental Biology, 34(2 Spec No), 345–351.
- Zheng, W., Wen, X., Zhang, B., & Qiu, Y. (2019). Selective effect and elimination of antibiotics in membrane bioreactor of urban wastewater treatment plant. The Science of the Total Environment, 646, 1293–1303. https://doi.org/10.1016/j.scitotenv.2018.07.400