ABSTRACT
One of the biggest hurdles of teaching data science and programming techniques to beginners is simply getting started with the technology. With multiple versions of the same coding language available (e.g., Python 2 and Python 3), various additional libraries and packages to install, as well as integrated development environments to navigate, the first step can be the most daunting. We show the advantages of using cloud computing to solve this issue and demonstrate one way of implementing it to allow beginners to get started with coding immediately. Using user-friendly Jupyter notebooks along with the interactive capabilities possible through Binder, we provide introductory Python and SQL material that students can access without downloading anything. This lets students to get started with coding right away without getting frustrated figuring out what to install. Example introductory modules on using Python and SQL for data analysis are provided through GitHub at https://github.com/Coleridge-Initiative/ada-intro-python and https://github.com/Coleridge-Initiative/ada-intro-sql.
1 Introduction
The big data revolution has resulted in a great need for new ways to teach data science tools. A large part of the improvements in data science education has been in revamping current curricula, introducing new data science courses as well establishing majors and degrees. Additionally, many tools have been developed and refined to facilitate traditional classroom learning of the modern data analytics workflow, such as incorporating version control with GitHub Classroom (Fiksel et al. Citation2019). In particular, the social sciences have lagged behind in keeping up with more modern statistical methods (Aiken et al. Citation1990; King Citation2011). Many efforts have been made to improve undergraduate and graduate education by targeting how the curricula are built to support data science (Nolan and Temple Lang Citation2010; Baumer Citation2015; Eybers and Hattingh Citation2016; Aikat et al. Citation2017; Cetinkaya-Rundel and Rundel Citation2018; Loy, Kuiper, and Chihara Citation2019; Yan and Davis Citation2019), and these efforts have extended to even secondary education (Gould et al. Citation2016).
However, the effort toward modernizing data analytics goes beyond the traditional classroom setting. Much of the focus has been on improving data science coursework and curricula, but an oft-overlooked segment of the population is working professionals such as staff at federal statistical agencies (Jarmin et al. Citation2014). Data science education is just as relevant for professionals who are already part of the workforce and have been working on important problems using restricted data already (Kreuter, Ghani, and Lane Citation2019). The challenges of building up professional development have been extensively discussed in a National Academies Roundtable on Data
Science Education (National Academies of Science, Engineering, and Medicine Citation2017). Employees at government agencies or researchers in the social sciences could also greatly benefit from learning these tools (Lane Citation2016). Part of the data science movement must also involve training working professionals, especially in the social sciences, where such efforts have been lacking (King Citation2011). While there are some options available for this audience in the academic class space, a more accessible method of learning data science is through workshops and short courses.
Though these forms of learning might be more convenient, they do come with their limitations. Instructor time is very limited in both workshops and online courses, and so the material must be very clear to be effective. Further, one unavoidable hurdle in these settings is the need to push through technological barriers associated with installing software. With a traditional 10–15 week academic course structure, students have more time and more contact with instructors to work out the kinks of the software they are working with. A computer lab or school library may even already be equipped with all the tools that are needed. These technological issues can become compounded in less traditional settings because of the condensed time frame. Alternative methods of delivering data science and programming material must be used in these cases.
In this article, we aim to demonstrate the ease with which cloud computing can be implemented to teach programming. First, in Section 2, we discuss the motivation behind using a cloud-based environment along with the benefits and drawbacks within the context of the type of course or program being offered. In this portion, we also provide a case study of using a cloud-based computing environment for a four-week online short course. Then, in Section 3, we provide a step-by-step guide for instructors to easily implement the methods described in the case study using Jupyter Notebooks, GitHub, and Binder. We present these tools as not only easy for students to use, but also for instructors to implement. Finally, we discuss where our method of using Jupyter and Binder fits in within the overall context of teaching with cloud computing in Section 4 and end with concluding remarks in Section 5.
2 Experiences Using a Cloud Computing Environment
2.1 Motivation
The Coleridge Initiative is a 501(c)(3) nonprofit developed to reach professionals in government agencies to improve their capabilities in utilizing their administrative data, with the goal of changing the empirical foundation of social science research. One of the main focuses of the Coleridge Initiative is to provide an Applied Data Analytics training program for government employees to build up the capacity of government agencies to use data science tools. The 6- or 10-day training programs cover a wide range of data analysis techniques, starting with basic descriptive statistics and building up to machine learning. Participants are also introduced to various related topics such as network analysis, text analysis, record linkage, and data privacy.
To make sure the in-person on-site class time was used as effectively as possible, the course material assumed a basic level of coding ability. Initially, participants were asked to complete material on DataCamp (https://www.datacamp.com), a self-paced online resource for learning introductory coding, but participants rarely used these due to the lack of interaction and structure (Kreuter, Ghani, and Lane Citation2019). To address this issue, an Introduction to Python and SQL online precourse was developed for participants to take leading up to the program. This four-week short course was designed to be taken mostly asynchronously with only weekly one-hour online video meetings as synchronous class time. Approximately 25 participants attended each of the synchronous sessions. The course covered an introduction to coding and data analysis in Python, along with running basic queries and doing joins in SQL. The goal of this course was to make sure that the in-class time would be spent on actually delving into data analysis rather than getting bogged down by coding issues.
There were a few key challenges associated with delivering such precourse material. First, the limited amount of synchronous time meant that the coding instructional material had to be as comprehensive as possible to allow for effective self-study. Software installation was another major hurdle, particularly when working with employees of government agencies. Adding any new piece of software to a government computer can be extremely time-consuming and frustrating. This problem is not limited to government employees though. Even with students in a traditional classroom, making sure the software is installed properly on each computer can be difficult. Differences between operating systems and inconsistencies in versions of software can lead to a variety of issues arising within even just a small class.
Finally, we needed to ensure that the material was as approachable as possible not only in terms of the difficulty of the material, but also in terms of the ease of use of the software. This was because many training program participants had no experience with any form of coding. Though some had done basic SQL queries before, many others had only Excel as the extent of their work with data analysis.
2.2 Cloud-Based Learning
Our goal was to develop a way to deliver an introductory Python and SQL course for data analysis that requires as little setup as possible for both instructors and students/participants. The key to achieving this goal came in the form of using a cloud-based learning environment. Cloud-based environments have been considered in various teaching applications for their ability to standardize hardware and software across users and their flexibility in scaling to demand (Cetinkaya-Rundel and Rundel Citation2018; National Academies of Sciences, Engineering, and Medicine 2018; Rexford et al. Citation2018). We considered several different options for the cloud-based instructional environment.
Two existing online environments were considered: Google Colab and RStudio Cloud. Google Colab, which is in the Google suite of products, offered an environment in which people could code and write plain text, similar to how one might use Google Docs except with additional coding capabilities (Bisong Citation2019). However, for our purposes, it did not offer enough flexibility of coding environments, particularly as we wanted to teach SQL without needing to run Python code. RStudio Cloud offered a user-friendly environment and an easy way to share workbooks using its integration with GitHub (Baumer et al. Citation2014; Cetinkaya-Rundel Citation2019), but required each user to create an account to access RStudio Cloud and limited us to working in the RStudio environment.
Using a JupyterHub server or a container-based method such as Docker was another option. There are many examples of using JupyterHub to share material for both collaborative and teaching purposes (Braun et al. Citation2017; Perkel Citation2018; Zonca and Sinkovits Citation2018; Kreuter, Ghani, and Lane Citation2019). Teaching data science with JupyterHub in particular has been well documented. The Berkeley National Workshop of Data Science Education has featured the use of JupyterHub for data science education (see https://data.berkeley.edu/external/workshops). The data science education team at University of California, Berkeley has been at the forefront of making these tools accessible to educators around the world (Adhikari and DeNero Citation2019). Wright et al. (Citation2019) discusses the usage of JupyterHub and other cloud environments for teaching computing in Biology classrooms.
The main downside was that setting up a JupyterHub server requires significant time, effort, and money on the part of the instructor (Holdgraf et al. Citation2017; Cetinkaya-Rundel and Rundel Citation2018). Though there is extensive documentation and instruction on setting up JupyterHub (Adhikari and DeNero Citation2019), it is still a daunting task for instructors without much experience in cloud environments. Setting up the server also requires buying cloud compute services if the instructor does not have access to space on a public-facing server. Our preference was for a free-to-use option over paid options.
The solution we arrived at came in the form of using Jupyter notebooks with Binder to deliver teaching material via a cloud-based environment (which we explain in greater detail in Section 3). The four week course consisted of four Jupyter notebooks that acted as class workbooks, with approximately 4 or 5 accompanying short (4–6 min) lecture videos each. The videos walked through specific concepts, such as using for loops or writing functions. These were supplemented with a one-hour synchronous virtual meeting in which the instructor would answer questions as well as walk through specific sections of the workbook while sharing their screen.
The cloud-based environment was set up so that students could access it anywhere, as long as they had internet access. They would simply click a link provided on the course website and, after a short wait, it would take them to an interactive workbook in which they could read descriptions, run examples, and write code. Afterward, they had the option of saving their work to their own computer. Importantly, the environment was fully browser-based and required zero installation or downloading (unless, of course, the students wanted to download the workbooks onto their own computers to save).
Despite running an online class for students who had never coded before and did not have any of the software (i.e., Python and SQLite) installed on their computers previously, we were able to run the class with few issues. The environment was user-friendly enough that everyone was able to easily access it, with the help of detailed guidelines provided on the course website. The set of Jupyter notebooks served as a combination of a textbook and a coding environment and all the reading and code-writing was done in the same place. Executable coding examples and built-in practice problems encouraged exactly this type of interaction. Students were able to jump right into running code from the very beginning, by executing the code already provided and then trying it out themselves within the same workbook. All code and output was displayed in the same window so there was not any need to render or build a notebook to see the code run. The success of the online introductory class allowed the Coleridge Initiative Applied Data Analytics program to move the basic Python and SQL material online before the in-person sessions, leaving more time for more advanced topics. It enabled participants to get up to speed on the prerequisite programming skills before arriving at the training program and made the most of a packed schedule.
In the next section, we demonstrate how to use Jupyter with Binder to easily provide access to interactive coding workbooks. The process of setting up Jupyter and Binder is designed to be as simple as possible so that instructors without much experience in utilizing cloud-based instructional tools can nevertheless feel comfortable with the process. There is of course more than one way to implement a cloud-based teaching environment. We provide this guide as an example of one method that is easy to learn and implement while highlighting the advantages that using a cloud-based environment has to offer.
3 Implementation of Cloud Computing for Teaching Using Jupyter and Binder
The most compelling argument for using Jupyter notebooks and Binder is that they can be used by instructors who might not consider themselves “tech-savvy.” The steps are:
Develop material using Jupyter notebooks.
Create a Git repository and fill the repo with notebooks as well as a configuration file.
Go to the Binder website (https://mybinder.org) and fill in the GitHub repository information.
In this section, we will walk through each step of the process and explain the tools used in each step. shows a rough overview of the process, broken down by the tools needed at each point. The most difficult part is in getting used to all of the new environments. Though it may seem daunting at first to try to combine various moving parts, we aim to demystify the process and show that anyone can use these tools. Examples shown in this article are generally constructed with Python, but instructors who have experience with working in a different programming language, such as R, can follow these steps to create and disseminate teaching material for that language as well.
Table 1 A summary of the tools and their uses for creating and delivering executable Jupyter notebooks.
3.1 Jupyter Notebooks
The Jupyter Project is a nonprofit organization developed to promote and facilitate open-source software development and reproducible research (Kluyver et al. Citation2016). This collaborative effort is focused primarily on the Jupyter Notebook, which is a coding environment that is able to seamlessly merge nicely formatted text with executable code written in any number of languages. The goal is to make code much more approachable for other researchers to view, understand, and run. Though primarily developed as a method for promoting collaborations between researchers, this same concept of accessible code can be applied to teaching new students (Smith Citation2016).
Juypter notebooks can be downloaded and installed using Python’s package manager, Pip. It may be easier for those unfamiliar with Pip to simply download and install Anaconda (https://www.anaconda.com/). Once downloaded and installed, the Anaconda launcher provides a nice graphical user interface (GUI) from which users can launch any number of tools in addition to Jupyter Notebooks (see ). Additional instructions about how to install Jupyter Notebooks can be found at https://jupyter.readthedocs.io/en/latest/install.html.
Jupyter notebooks are viewed in a browser and look similar to a Google or Word document. The main difference is that the writing is done in individual cells that can be toggled to be used for code or for writing narrative text. shows an example of a section in a Jupyter notebook containing a code cell surrounded by explanations outside of the coding syntax. The combination of code and plain text is much more inviting than asking new coding students to start with a command line or text editor. Once inside a new Jupyter Notebook environment, users may start writing in the document within individual cells.
Fig. 2 Jupyter notebook cells allow for code blocks to be integrated into the descriptions. There are three cells in this section: the top cell contains information about loading libraries, the middle cell runs some code, and the bottom cell provides additional explanations.

There are two main types of cells that will account for the vast majority of use cases in Jupyter notebooks: code cells and Markdown cells. Code cells allow users to run code, with the output of the code appearing directly below the cell. Each notebook has an associated kernel. A kernel is a computing process that allows users to specify which programming language to use in the code cells. To create a Python notebook, the user would simply choose the Python kernel for that notebook. Users may run the code cells by using Shift + Enter (which runs the cell and takes the cursor to the next cell) or Ctrl + Enter (which runs the cell and keeps the cursor in the same cell), or by clicking the Run button in the toolbar. The output is displayed directly below that code cell. Only the code within the cells will be run, instead of the entire document, so that code can be executed a few lines at a time. This has the added benefit of allowing students to run code step by step to debug their code and identify where they might have made a mistake.
Markdown cells allow users to write narrative text to accompany the code. Markdown is a lightweight formatting markup language with easy-to-use rules to render text. Though students may not be familiar with using Markdown formatting for text, it is quite often quick to pick up. There are very simple rules to make documents nicer to read, such as using headers (using the # symbol, with multiple denoting sub-headers), italics (using * on either side of text to be italicized), or boldface (using ** on either side). Running the cells as mentioned above will render the Markdown formatting. Though students will make less use of this type of cell, it is nevertheless crucial in making Jupyter notebooks an effective teaching tool. The Markdown cells allow instructors to insert descriptions and even include images or tables directly in the document that students code in without having to resort to comments or adding unwieldy extra code. The combination of the code cells with the Markdown cells not only makes the entire coding process approachable for students, it also allows for instructors to include guides and examples alongside questions and practice problems for students to solve.
Adding additional cells is done using the “+” button on the toolbar at the top of the document. Additional commands allow users to cut, copy, paste, rearrange, split, or merge entire cells. Output cells may be hidden and collapsed by clicking on a space to the left of each output. Various keyboard shortcuts exist for each of these operations. As users get more comfortable with the environment, navigating the notebook can become quite smooth. Notably, the entire notebook does not need to be run to view and render Markdown formatting. Individual code and Markdown cells may be run independently of each other for a more pleasant viewing experience. To avoid problems that can arise from running cells out of order, we do suggest making the cells as independent and self-contained as possible outside of the initial loading of packages and data.
Though initially designed to be used with a few different programming languages—namely, Julia, Python, and R, providing the origins of its name—Jupyter now supports a wide array of languages through a number of community-contributed kernels (see https://github.com/jupyter/jupyter/wiki/Jupyter-kernels for a list of supported kernels). This means that Jupyter notebooks are a highly flexible teaching tool able to support many different types of instructional programs.
3.2 GitHub
The next step in the process is to put everything in a GitHub repository. Git and GitHub have been the subject of many pedagogical discussions (Lawrance, Jung, and Wiseman Citation2013; Haaranen and Lehtinen Citation2015; Fiksel et al. Citation2019). Though there are many advantages to using Git—particularly version control—for the purposes of delivering executable notebooks, we do not actually need to use the full capabilities of the Git repository. Instead, we simply use it as a form of file storage space.
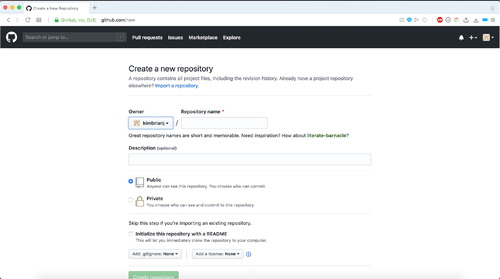
To start, you can go to https://github.com and create a free account. shows the information that needs to be filled out to create a new GitHub repository. Simply provide a descriptive name and create the new repository. You may also choose to include a license at this point, choosing one from the dropdown menu near the bottom of the page. This will automatically include a LICENSE.md file in your repository as it is created. Licensing your repository can make it so that it is open source if you wish and encourage sharing and reproduction of these teaching tools. For the examples provided in this article, we have used a Creative Commons Zero v1.0 Universal license, which waives copyright interest and makes it available to anyone who wishes to use it. More information on licensing on GitHub can be found at https://docs.github.com/en/free-pro-team@latest/github/creating-cloning-and-archiving-repositories/licensing-a-repository.
Once the repository has been created, Jupyter notebooks can now be uploaded from your local computer to GitHub by clicking on the “Upload” button in the newly-made repository. Any data used in the notebooks, such as CSV files, should also be uploaded. A separate folder for data is not necessary, though it is recommended when there are many data files. GitHub has a strict 100 MB file size limit, so larger data files will need to be loaded in the notebooks through another method, such as reading them in from a URL.
The last file to add is a configuration file. The configuration file allows you to specify packages that you want to include and have installed in the environment by including a plain text file with the proper information. The file is formatted either as an environment.yml file (a .yml file is simply a plain text file used to specify the configuration) or a requirements.txt file. Conda (Anaconda’s package management tool) is used to install the packages if an environment.yml is the one included. shows an example of an environment.yml file, with the packages listed under dependencies. The only text that might change would be the “name” filed, indicating the name of the repository, and the “dependencies” field, indicating a list of packages to be installed. The “channels” field refers to how the packages will be installed and should not be changed.
Fig. 4 The environment.yml and requirements.txt files are quite straightforward—simply fill in the name and change the list of dependencies to match whatever packages you want to use.

If the notebooks are limited to only Python code, users may opt for the simpler requirements.txt file. This plain text file will just need to list the packages to be installed, with each package on its own line. The packages are installed using pip. shows an example of a requirements.txt file. More information on how to construct the configuration file can be found at https://mybinder.readthedocs.io/en/latest/config_files.html.
While we ignore Git’s functionalities here, more advanced courses or programs may elect to provide an overview of how Git works and encourage students to pull the necessary materials directly from a class repository, as encouraged by Fiksel et al. (Citation2019). We do note that this is not a trivial step, due in part to the JSON format of Jupyter notebooks making merging more complicated and in part due to lack of a convenient Git pane in Jupyter.
3.3 Binder
The last step to deploying the Jupyter notebooks is to use Binder. Binder is a free-to-use online tool to easily create a cloud-computing environment using a GitHub repository. It was initially developed for sharing reproducible code to improve collaborative efforts between researchers (Project Jupyter et al. Citation2018). It was designed to make sharing executable code easy so that anyone who was new to the code could open and run the code without needing to worry about installation of packages or dependencies. Here, we take this same idea and apply it to simplifying the coding setup for students.
Binder works by creating an interactive computational environment using Kubernetes and JupyterHub, setting up a cloud computing environment automatically for anyone who wants to use it (Project Jupyter et al. Citation2018). Though the cloud computing setup would normally require some amount of technical expertise and knowledge of how tools like Docker and JupyterHub work, the Binder website (https://mybinder.org) automates this process. Using Binder allows instructors to use these tools without requiring any technical knowledge. As long as the GitHub repository is set up correctly, you can simply go to the website, fill in the GitHub repository URL, and then click the “Launch” button. This will generate a URL that you can use to launch Binder with the contents of your GitHub repository. shows the Binder website with the fields that can be filled in.
Fig. 5 After putting the Jupyter notebooks on GitHub, setting them up with Binder can be accomplished in a few small steps.
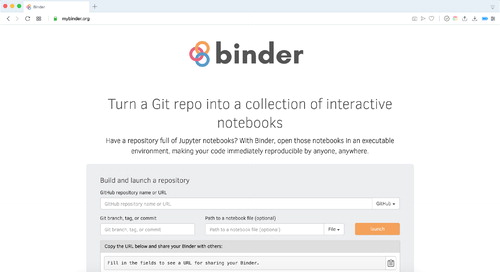
The URL provided on the site allows you to launch the environment to run the Jupyter notebooks. The first time you launch Binder for your notebooks, it will take a while to build everything. We recommend instructors launch Binder before deploying it so that they can both make sure that opening it takes as little time as possible for students and participants.
While Binder is offered for free through their website, it does have a few limitations. First, users are limited to between 1 GB and 2 GB of RAM. In our applications, since we were teaching basic concepts in Python and SQL, we found that we never ran into any issues with the amount of RAM allotted to the user sessions. Second, Binder sessions time out after 10 min of inactivity to conserve resources. In practice, we found that though this was a possible source of annoyance for students, we were able to work around it. As long as we warned them about this limitation and provided them with strategies for avoiding the issues (e.g., encouraging them to save work locally before moving on), students were able to work around the limitations. Finally, the work that students do in the environment did not persist over multiple sessions, since Binder does not offer the option of saving work on their servers. We encouraged students to save work locally to keep their work, which was easily done through Jupyter.
The GitHub repositories containing the introductory Python and SQL material we used for the Applied Data Analytics training program can be found at https://github.com/Coleridge-Initiative/ada-intro-python and https://github.com/Coleridge-Initiative/ada-intro-sql, and the interactive environment can be accessed by clicking the “Launch Binder” button on the GitHub page. We have also used the Binder environment to deliver a half-day module on pulling from the PatentsView API, and the notebook and Binder link for it can be found at https://github.com/Coleridge-Initiative/patentview-api.
4 Discussion
We have provided a guide for using Jupyter and Binder to teach courses, mainly discussing its uses in teaching Python. However, Jupyter Notebooks are extremely flexible and can be used with a wide variety of languages. The steps outlined here will be widely applicable, but some languages will be easier than others. For example, adapting these steps to use R rather than Python will be as easy as replacing a Python kernel notebook with an R kernel notebook (and putting in the appropriate R code).
Others may take a little more work. We have also used these tools to teach introductory SQL, which requires a database set up to practice any coding. Using the BeakerX collection of kernels and extensions to Jupyter notebooks, we were able to develop SQL notebooks that ran queries and displayed the results in the notebook. Since teaching SQL required a database to query from, we had to include a script that unzips compressed data files and creates the database as Binder is launched. An example of this script can be found at https://github.com/Coleridge-Initiative/ada-intro-sql/blob/master/binder/postBuild.
Though motivated initially by the Applied Data Analytics training programs, using Jupyter and Binder to address these challenges is applicable in a wide variety of settings. Many instructors are moving teaching material online so that they can reach more people. Even without the strict security restrictions imposed by government agencies, some may find that their computers are not able to install and run the software necessary to learn from such online courses. The cloud environment is accessed through a browser, so the notebooks work regardless of the operating system the user is running, a benefit that is useful in all contexts.
Instructors may find this method useful even for technical audiences for short courses and demonstrations. Since there is relatively little time during short courses, trying to get everyone set up with the appropriate packages and versions can present a real barrier. By the time all the individual issues are sorted out, the course or workshop may already be over. Instructors can instead make sure that the appropriate packages are already installed (i.e., the requirements file contains the correct information) and be certain that everyone will be using the same version and same packages.
Though initially developed for training programs outside of a traditional university setting, the tools presented here can be used for those classes as well. A four-week online course called Introduction to Python and SQL (SURV 672) has been run using Binder as the primary method of programming at University of Maryland College Park, using much of the same structure as in the class run for government agencies through the Coleridge Initiative.
There are downsides to using cloud-based environments for learning. Internet access is necessary to do any coursework, which can be inconvenient in some cases. Even if the student does have internet access, it needs to be stable enough to work for an extended period of time. Though the restrictions are not high, as the computational burden is not on the students’ machines, a spotty connection can be a source of frustration as students may lose progress or be unable to run code without knowing why.
In our particular implementation, students were also unable to save work in the cloud despite doing all of their work in the cloud, requiring them to instead download all work and upload every time they wanted to continue working on the same workbook. We worked around this issue by making it clear that the notebooks were intended as a place to try out writing a few lines of code, rather than using them for full coding tasks. Students generally did not have any complaints about saving their work and were mostly only interested in downloading the notebook at the end of a session to view later, rather than carrying over any partially completed work.
We found that though these issues did pop up, they were relatively infrequent and easily addressable. For example, unlike software installation issues, internet problems tended to be more temporary and were more of an annoyance rather than a full roadblock. We also tried to keep workbooks short enough that they could be completed in relatively few chunks (or even in one sitting), minimizing the need to save notebooks. In general, most participants reported a smooth experience and were able to access and use the workbooks without much trouble.
One possible area of improvement is the implementation of assessment tools and techniques. The only assessment that took place in our short course was in the form of questions and answers during the one-hour synchronous meetings. This was because we avoided larger coding tasks, due to the inability to save. We also wanted to avoid possible pitfalls on our end when delivering the material, so we intentionally tried to keep the complexity of the notebooks down. We provided solutions to the simple questions asked in each notebook, but no tools such as a Jupyter extension called nbgrader were implemented. Future extensions of using the methods described in this article would include more extensive application of these assessment tools. Though some of the JupyterHub integration would be lost when implementing nbgrader through Binder, the robust code-checking provided by nbgrader would be a potentially huge benefit (Project Jupyter et al. Citation2019).
Other extensions include combining all of the Jupyter Notebooks into a Jupyter Book. This would provide a way for instructors to include “Interact” buttons at the top of each section, allowing students to work on individual notebooks one at a time. The Data 8: Fundamentals of Data Science course at University of Berkeley has used Jupyter Book for its course textbook (Adhikari and DeNero Citation2019). Similarly, even if Binder is not used as a main programming environment in traditional courses, instructors may choose to make additional or optional code available on their GitHub pages and simply add a Binder link for students wishing to jump right in and replicate what the instructor did.
5 Conclusion
We have presented an easy-to-implement method for delivering interactive programming instructional material without requiring students to install anything on their own computers. We encourage instructors to consider cloud computing environments in general for their teaching activities. In our experience, Jupyter and Binder are both fast to set up by instructors and intuitive to follow for students, making them ideal for training programs and workshops where time is limited and easily accessible material is essential.
ORCID
Brian Kim http://orcid.org/0000-0002-2278-2432
References
- Adhikari, A., and DeNero, J. (2019), “Computational and Inferential Thinking: The Foundations of Data Science,” GitBook, licensed under CC BY-NC-ND 4.0.
- Aikat, J., Carsey, T. M., Fecho, K., Jeffay, K., Krishnamurthy, A., Mucha, P. J., Rajasekar, A., and Ahalt, S. C. (2017), “Scientific Training in the Era of Big Data: A New Pedagogy for Graduate Education,” Big Data, 5, 12–18. DOI: 10.1089/big.2016.0014.
- Aiken, L. S., West, S. G., Sechrest, L., Reno, R. R., Roediger, III, H. L., Scarr, S., Kazdin, A. E., and Sherman, S. J. (1990), “Graduate Training in Statistics, Methodology, and Measurement in Psychology: A Survey of PhD Programs in North America,” American Psychologist, 45, 721–734. DOI: 10.1037/0003-066X.45.6.721.
- Baumer, B. (2015), “A Data Science Course for Undergraduates: Thinking With Data,” The American Statistician, 69, 334–342. DOI: 10.1080/00031305.2015.1081105.
- Baumer, B., Cetinkaya-Rundel, M., Bray, A., Loi, L., and Horton, N. J. (2014), “R Markdown: Integrating A Reproducible Analysis Tool Into Introductory Statistics,” Technological Innovations in Statistics Education, 8. Available at https://escholarship.org/uc/item/90b2f5xh.
- Bisong, E. (2019), “Google Colaboratory,” in Building Machine Learning and Deep Learning Models on Google Cloud Platform, Berkeley, CA: Apress.
- Braun, N., Hauth, T., Pulvermacher, C., and Ritter, M. (2017), “An Interactive and Comprehensive Working Environment for High-Energy Physics Software With Python and Jupyter Notebooks,” Journal of Physics: Conference Series, 898, 072020. DOI: 10.1088/1742-6596/898/7/072020.
- Cetinkaya-Rundel, M. (2019), “Computing Infrastructure and Curriculum Design for Introductory Data Science,” in Proceedings of the 50th ACM Technical Symposium on Computer Science Education, SIGCSE ’19, Association for Computing Machinery, New York, NY, USA, p. 1236.
- Cetinkaya-Rundel, M., and Rundel, C. (2018), “Infrastructure and Tools for Teaching Computing Throughout the Statistical Curriculum,” The American Statistician, 72, 58–65. DOI: 10.1080/00031305.2017.1397549.
- Eybers, S., and Hattingh, M. (2016), “Teaching Data Science to Post Graduate Students: A Preliminary Study Using a ‘F-L-I-P’ Class Room Approach,” in International Association for Development of the Information Society.
- Fiksel, J., Jager, L. R., Hardin, J. S., and Taub, M. A. (2019), “Using GitHub Classroom to Teach Statistics,” Journal of Statistics Education, 27, 110–119. DOI: 10.1080/10691898.2019.1617089.
- Gould, R., Machado, S., Ong, C., Johnson, T., Molyneux, J., Nolen, S., Tangmunarunkit, H., Trusela, L., and Zanontian, L. (2016), “Teaching Data Science to Secondary Students: The Mobilize Introduction to Data Science Curriculum,” in Promoting Understanding of Statistics about Society, eds. J. Engel. Proceedings of the Roundtable Conference of the International Association of Statistics Education (IASE), July 2016, Berlin, Germany.
- Haaranen, L., and Lehtinen, T. (2015), “Teaching Git on the Side-Version Control System as a Course Platform,” in Proceedings of the 2015 ACM Conference on Innovation and Technology in Computer Science Education.
- Holdgraf, C., Culich, A., Rokem, A., Deniz, F., Alegro, M., and Ushizima, D. (2017), “Portable Learning Environments for Hands-On Computational Instruction,” in Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact. DOI: 10.1145/3093338.3093370.
- Jarmin, R., Lane, J., Marco, A., and Foster, I. (2014), “Using the Classroom to Bring Big Data to Statistical Agencies,” AMSTAT News: The Membership Magazine of the American Statistical Association, 449, 12–13.
- King, G. (2011), “Ensuring the Data-Rich Future of the Social Sciences,” Science, 331, 719–721. DOI: 10.1126/science.1197872.
- Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B., Bussonnier, M., Frederic, J., Kelley, K., Hamrick, J., Grout, J., Corlay, S., Ivanov, P., Avila, D., Abdalla, S., and Willing, C. (2016), “Jupyter Notebooks—A Publishing Format for Reproducible Computational Workflows,” in Positioning and Power in Academic Publishing: Players, Agents and Agendas, eds. F. Loizides and B. Schmidt, Amsterdam: IOS Press, pp. 87–90.
- Kreuter, F., Ghani, R., and Lane, J. (2019), “Change Through Data: A Data Analytics Training Program for Government Employees,” Harvard Data Science Review, 1. DOI: 10.1162/99608f92.ed353ae3.
- Lane, J. (2016), “Big Data for Public Policy: The Quadruple Helix,” Journal of Policy Analysis and Management, 35, 708–715. DOI: 10.1002/pam.21921.
- Lawrance, J., Jung, S., and Wiseman, C. (2013), “Git on the Cloud in the Classroom,” in Proceeding of the 44th ACM Technical Symposium on Computer Science Education, pp. 639–644. DOI: 10.1145/2445196.2445386.
- Loy, A., Kuiper, S., and Chihara, L. (2019), “Supporting Data Science in the Statistics Curriculum,” Journal of Statistics Education, 27, 2–11. DOI: 10.1080/10691898.2018.1564638.
- National Academies of Science, Engineering, and Medicine (2017), Roundtable on Data Science Postsecondary Education Meeting #4, available at https://www.nationalacademies.org/event/10-20-2017/roundtable-on-data-science-postsecondary-education-meeting-4.
- National Academies of Science, Engineering, and Medicine (2018), Data Science for Undergraduates: Opportunities and Options, Washington, DC: The National Academies Press.
- Nolan, D., and Temple Lang, D. (2010), “Computing in the Statistics Curricula,” The American Statistician, 64, 97–107. DOI: 10.1198/tast.2010.09132.
- Perkel, J. M. (2018), “Why Jupyter Is Data Scientists’ Computational Notebook of Choice,” Nature, 563, 145–146. DOI: 10.1038/d41586-018-07196-1.
- Project Jupyter, Blank, D., Bourgin, D., Brown, A., Bussonnier, M., Frederic, J., Granger, B., Griffiths, T. L., Hamrick, J., Kelley, K., Pacer, M., Page, L., Pérez, F., Ragan-Kelley, B., Suchow, J. W., and Willing, C. (2019), “nbgrader: A Tool for Creating and Grading Assignments in the Jupyter Notebook,” The Journal of Open Science Education, 2, 32. DOI: 10.21105/jose.00032.
- Project Jupyter, Bussonnier, M., Forde, J., Freeman, J., Granger, B., Head, T., Holdgraf, C., Kelley, K., Nalvarte, G., Osheroff, A., Pacer, M., Panda, Y., Perez, F., Ragan-Kelley, B., and Willing, C. (2018), “Binder 2.0—Reproducible, Interactive, Sharable Environments for Science at Scale,” in Proceedings of the 17th Python in Science Conference. DOI: 10.25080/Majora-4af1f417-011.
- Rexford, J., Balazinska, M., Culler, D., and Wing, J. (2018), “Enabling Computer and Information Science and Engineering Research and Education in the Cloud,” Tech. Rep.
- Smith, A. A. (2016), “Teaching Computer Science to Biologists and Chemists, Using Jupyter Notebooks: Tutorial Presentation,” Journal of Computing Sciences in Colleges, 32, 126–128.
- Wright, A. M., Schwartz, R. S., Oaks, J. R., Newman, C. E., and Flanagan, S. P. (2019), “The Why, When, and How of Computing in Biology Classrooms,” F1000Research, 8, 1854. DOI: 10.12688/f1000research.20873.1.
- Yan, D., and Davis, G. E. (2019), “A First Course in Data Science,” Journal of Statistics Education, 27, 99–109. DOI: 10.1080/10691898.2019.1623136.
- Zonca, A., and Sinkovits, R. S. (2018), “Deploying Jupyter Notebooks at Scale on XSEDE Resources for Science Gateways and Workshops,” in Proceedings of the Practice and Experience on Advanced Research Computing, PEARC ’18, Association for Computing Machinery, New York, NY, USA. DOI: 10.1145/3219104.3219122.