 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
This study presents a separable nonlinear least squares (SNLLS) implementation of the minimum distance (MD) estimator employing a fixed-weight matrix for estimating structural equation models (SEMs). In contrast to the standard implementation of the MD estimator, in which the complete set of parameters is estimated using nonlinear optimization, the SNLLS implementation allows a subset of parameters to be estimated using (linear) least squares (LS). The SNLLS implementation possesses a number of benefits, such as faster convergence, better performance in ill-conditioned estimation problems, and fewer required starting values. The present work demonstrates that SNLLS, when applied to SEM estimation problems, significantly reduces the estimation time. Reduced estimation time makes SNLLS particularly useful in applications involving some form of resampling, such as simulation and bootstrapping.
1. Introduction
This study addresses the application of separable nonlinear least squares (SNLLS) when performing covariance structure analysis (CSA). SNLLS was first introduced by Golub and Pereyra (Citation1973), who showed that for a certain type of nonlinear estimation problems, a subset of parameters can be estimated using numerically efficient least squares (LS). As will be discussed below, several studies have shown that parameter separation offers a number of numerical benefits, such as faster convergence, better performance when the estimation problem is ill-conditioned (i.e., problems in which the ratio between the largest and the smallest singular value of the covariance matrix is large), and fewer required starting values.
SNLLS is typically applied to problems involving some form of nonlinear regression analysis, but not exclusively so. A recent study by Kreiberg et al. (Citation2021) suggested an SNLLS implementation of the minimum distance (MD) estimator for estimating confirmatory factor analysis (CFA) models. The motivation for the current study is to generalize the results in Kreiberg et al. (Citation2021) by outlining an SNLLS implementation for estimating structural equation models (SEMs). This is important for several reasons. First, it makes SNLLS applicable to a wider range of models. Second, at this stage, little is known about the potential benefits of applying SNLLS in the context of CSA. The outlined SNLLS implementation may pave the way for future research on how to improve the numerical performance of CSA based estimators.
To make the idea of SNLLS clearer, consider the familiar MD quadratic form objective function
(1)
(1)
where
and
are covariance vectors derived from the sample and the model, respectively,
is the parameter vector and
is a weighting matrix chosen by the user. We consider the case in which
is a fixed matrix (i.e., when
is not a function of
). Such cases include well-known estimators such as unweighted least squares (ULS), generalized least squares (GLS), and weighted least squares (WLS). The standard implementation of EquationEquation (1)
(1)
(1) is a one-step estimation procedure, here referred to as nonlinear least squares (NLLS), that involves the use of nonlinear optimization techniques. Estimation is performed by searching the parameter space for the value of
that minimizes EquationEquation (1)
(1)
(1) . In contrast, the SNLLS implementation of EquationEquation (1)
(1)
(1) is a two-step estimation procedure that works by splitting
into two subsets. In the first step, one subset of parameters is estimated using nonlinear optimization. In the second step, based on the estimates obtained in the first step, the remaining subset of parameters is estimated using LS. As demonstrated in Kreiberg et al. (Citation2021), SNLLS provides parameter estimates and a minimum objective function value identical to those obtained using NLLS. It obviously follows that the asymptotic properties of the estimator are maintained. The presentation below presents a general framework for how to accomplish parameter separation in the case of SEMs.
Over the years, SNLLS has become popular in applied research across a wide range of scientific disciplines. Golub and Pereyra (Citation2003) compiled a list of real-world examples of SNLLS applications. Mullen (Citation2008) subsequently provided a comprehensive overview of SNLLS for a number of applications in physics and chemistry. SNLLS has also proved useful in systems and control applications. For instance, Söderström et al. (Citation2009), Söderström & Mossberg (Citation2011), and Kreiberg et al. (Citation2016) applied CSA to handle the errors-in-variables (EIV) estimation problem. The work in these studies showed how to implement the MD estimator using SNLLS.
Several studies have documented that the SNLLS implementation of nonlinear estimators offers a number of benefits. For instance, Sjöberg and Viberg (Citation1997) evaluated the numerical performance of SNLLS when applied to neural-network minimization problems. Their main conclusions were that SNLLS provides faster convergence and performs better in cases in which the estimation problem is ill-conditioned. A recent study by Dattner et al. (Citation2020) investigated the performance of SNLLS when applied to estimation problems involving ordinary differential equations (ODEs). Their simulations showed that SNLLS provides faster convergence as well as parameter estimates of similar or higher accuracy than what is achieved by traditional nonlinear procedures.
The remainder of this article is organized as follows. Section 2 establishes the notation used throughout the article. In this section, we provide a brief overview of the SEM framework and the associated MD estimator. Section 3 outlines how to modify the MD objective function to accommodate the SNLLS implementation of the estimator when applied to SEMs. Section 4 compares the numerical efficiency of SNLLS and NLLS when applied to real-world estimation problems. Finally, Section 5 presents some concluding remarks.
2. Background
2.1. Notation
Before presenting the SEM framework, it will be useful to introduce the following notation. Let be a
zero-mean random vector, and let
be the associated
covariance matrix given by
(2)
(2)
where
is the expectation operator and the superscript T is the transpose of a vector or a matrix. The number of nonredundant elements in
is
given that no restrictions other than symmetry are placed on the elements of
A covariance vector containing the nonredundant elements (i.e., the lower half of
including the diagonal) is
(3)
(3)
In this expression,
is the operation of vectorizing the nonredundant elements of
Alternatively,
is obtained by
(4)
(4)
Here,
is the operation of vectorizing the elements of a matrix by stacking its columns, and
is a
matrix obtained from
(5)
(5)
where
is a
selection matrix containing only ones and zeros. This matrix has the additional usage
(6)
(6)
In the case of symmetry,
is referred to as the duplication matrix in the literature (see Magnus & Neudecker, Citation1999). The matrices
and
can be formed to handle covariance matrices with additional structure beyond symmetry. For instance, in the case that
is a diagonal,
is constructed so that
contains only the elements on the diagonal of
Appendix A outlines a general framework for how to obtain
and
for various structures characterizing
We now expand the previous notation. Let and
be
and
zero-mean random vectors, respectively. A
dimensional column vector is given by
(7)
(7)
The associated
covariance matrix is
(8)
(8)
where
(9)
(9)
As before, the vector consisting of the nonredundant elements of is given by
However, for later, it will be more convenient to work with the vector
(10)
(10)
where
(11)
(11)
Note that contains the same elements as
but in a different order. The last equation in EquationEquation (11)
(11)
(11) follows from the fact that there is no redundancy in
Appendix A shows how to derive a matrix
Then, by using EquationEquations (4)
(4)
(4) and Equation(5)
(5)
(5) , we obtain the covariance vector
(12)
(12)
2.2. The SEM Framework
With the basic notation in place, we are ready to introduce the SEM framework, which consists of the following three equations (excluding constant terms)
(13)
(13)
(14)
(14)
(15)
(15)
The first equation is the structural equation, which specifies the causal relationships among the latent variables. In this equation,
and
are respectively
and
random vectors,
is a
random noise vector, and
and
are respectively
and
parameter matrices relating the latent random vectors. The last two equations are measurement equations. In these equations,
and
are respectively
and
observed random vectors,
and
are noise vectors of similar dimensions, and
and
are respectively
and
parameter matrices relating the observed and the latent random vectors. All random vectors are zero-mean.
It is assumed that , where I is the identity matrix, is nonsingular such that
is uniquely determined by
and
It is further assumed that
and
are mutually uncorrelated, and that
and
are mutually uncorrelated with
and
respectively. The noise vectors
and
are allowed to correlate.
The specification additionally includes the following covariance matrices
(16)
(16)
The nonredundant elements of
and
are given by the covariance vectors
(17)
(17)
Let
be a parameter vector containing the free elements in
and
and let
The covariance matrix implied by EquationEquations (13)–(15) is
(18)
(18)
2.3. The MD Estimator
Suppose that a sample of data points (for
) is available. An estimate of
is then computed using
(19)
(19)
Given
the aim is to estimate the true parameter vector
An estimate of
is obtained by
(20)
(20)
where
is a scalar function that expresses the distance between the observed and the model-implied covariance structure. Below, we focus on the MD objective function given by
(21)
(21)
In this expression,
and
are vectors containing the nonredundant elements of
and
respectively. That is,
(22)
(22)
Moreover, the matrix
is a positive definite weighting matrix. Under suitable conditions, and for the right choice of
the MD estimator is consistent and asymptotically normal. Note that consistency does not depend on
as long as
converges in probability to a symmetric positive definite matrix.
Using a proper algorithm, EquationEquation (21)(21)
(21) is minimized by numerically searching the parameter space until some convergence criterion is satisfied. For the estimation problem to be feasible, it is a necessary condition that the number of elements in
is at least as large as the number of free parameters in
3. Modifying the MD Quadratic Form Objective Function
Next, we outline how to modify the objective function in EquationEquation (21)(21)
(21) to accommodate the SNLLS implementation. To do so, we need some additional notation. Let
be a
vector containing the free elements in
, and
and let
be a
vector containing the free elements in
, and
The vector
is formed by
(23)
(23)
The complete parameter vector now becomes
(24)
(24)
The key to applying SNLLS is the separation of parameters, which involves expressing the covariance vector in EquationEquation (10)
(10)
(10) using
(25)
(25)
In this expression,
is a tall matrix valued function (i.e., a matrix consisting of more rows than columns) assumed to have full column rank. In Appendix B, it is shown that
takes the general form
(26)
(26)
where
is the Kronecker product, the 0s are zero matrices of compatible sizes, and
is the identity matrix. It is now possible to write the objective function using
(27)
(27)
where
and
correspond to
and
respectively, but with their rows and columns rearranged according to the order in
For some value of
the solution to the problem of minimizing EquationEquation (27)
(27)
(27) w.r.t.
is a straightforward application of LS
(28)
(28)
Since
depends on
, it is necessary to outline how to obtain an estimate
without directly involving
. Theorem 2.1 in Golub and Pereyra (1973) provides the justification for replacing
in EquationEquation (27)
(27)
(27) with the right-hand side of EquationEquation (28)
(28)
(28) . Doing so, leads to the modified objective function
(29)
(29)
Apart from some slight notational differences, the derivation of EquationEquation (29)
(29)
(29) is similar to the derivation in Kreiberg et al. (Citation2021). From the preceding presentation, it follows that SNLLS is a two-step procedure. In the first step,
is obtained by minimizing EquationEquation (29)
(29)
(29) applying nonlinear optimization. In the second step, using
from the first step,
is obtained by EquationEquation (28)
(28)
(28) .
The major benefit of the formulation in EquationEquation (29)(29)
(29) is that the minimization w.r.t.
represents a lower dimensional optimization problem. Thus, the computational load when minimizing
w.r.t.
is smaller, and in some cases by a considerable margin, than what is the case when minimizing EquationEquation (21)
(21)
(21) w.r.t.
This is especially the case when the number of elements in
is large compared with the number of elements in
4. Illustrations
This section provides two examples that illustrate the difference in numerical efficiency between the two implementations, SNLLS and NLLS, of the MD estimator when applied to SEMs. Numerical performance is assessed by studying the convergence of the optimizer and the time it takes the optimizer to reach its minimum. Since timing depends on other processes running on the device performing the estimation, it is recommended to compute the average estimation time over multiple runs. Estimation and timing are performed using Matlab (Citation2020, version R2020b). The two implementations are compared under the following conditions:
Algorithm: The optimizer is a Quasi-Newton (QN) design applying the Broyden–Fletcher–Goldfarb–Shanno (BFGS) Hessian update mechanism (default in Matlab).
Gradient: For simplicity, the gradient is computed using a finite difference approach. The computation is based on a centered design, which is supposed to provide greater accuracy at the expense of being more time-consuming.
Tolerances: Tolerances are set to their default values (details are found in the Matlab documentation).
Starting values: Starting values are taken from the open-source R (R Core Team, Citation2021) package lavaan (Rosseel, Citation2012). The starting values for the free elements are as follows:
–
and
are computed using the non-iterative fabin 3 estimator (see Hägglund, Citation1982).
–
and
are set to zero.
–
and
are set to zero except for the diagonal elements, which are set to 0.05.
–
and
are set to zero except for the diagonal elements, which are set to half the observed variance. For the examples below, no starting values are required for the elements in
,ϵ1
Estimator: The GLS estimator is used throughout the examples. The GLS estimator uses a weight matrix of the form
(30)
(30)
Timing: In each example, the model is re-estimated 1000 times using the same empirical covariance matrix as input.
4.1. Example 1
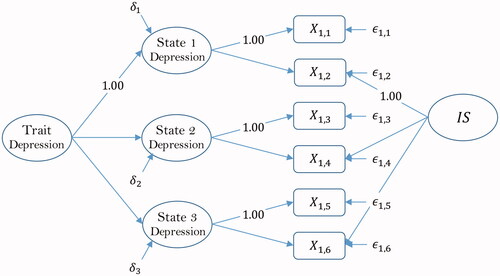
The first example considers a model for the medical illness of depression. The data ( used in this example are taken from Geiser (Citation2012) and consist of six indicators of depression. In the data,
and
are indicators of the first-order common factor Depression State 1,
and
are indicators of the first-order common factor Depression State 2, and
and
are indicators of the first-order common factor Depression State 3. The three factors themselves are indicators of the second-order common trait factor Depression. The model additionally contains an indicator-specific factor labeled IS. Indicators
, and the factor Depression State 1 serve as marker variables. The path diagram illustrating the structure of the model is shown in .
Figure 1. Geiser (Citation2012).
Results of the estimation are presented in . As seen from the table, the number of iterations and function evaluations is (It, Fe) (23, 375) for SNLLS and (It, Fe)
(145, 5439) for NLLS. As expected, the required computational load for minimizing
w.r.t.
is far less than the required load for minimizing
w.r.t.
shows the convergence profiles for the two implementations. From the figure, it is clear that the SNLLS objective function
starts at a point much closer to its minimum of 0.0109 than what is seen for the NLLS objective function
In terms of estimation time, the mean time is 0.0334 sec. for SNLLS and 0.1408 sec. for NLLS. Thus, SNLLS is faster by a factor of 0.1408/0.0334
4.2153. The results in this example clearly suggest that the SNLLS implementation is numerically more efficient than the standard NLLS implementation.
Figure 2. Convergence profile, Geiser (Citation2012).
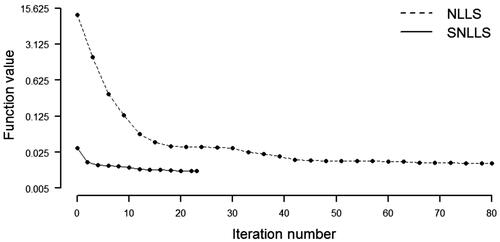
Table 1. Timing results, Geiser (Citation2012).
4.2. Example 2
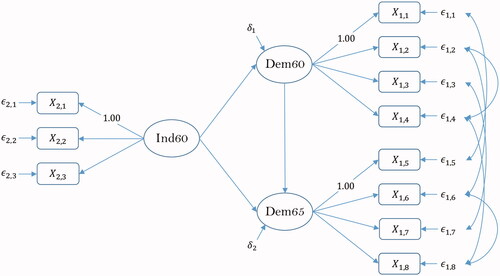
The second example considers a model for industrialization and political democracy. The model is taken from Bollen (Citation1989), and has been used extensively in books, tutorials, etc. The data consist of 11 indicators of industrialization and political democracy for 75 countries ( In the data,
are indicators of the common factor Political Democracy at time 1 (1960),
are indicators of the common factor Political Democracy at time 2 (1965) and
are indicators of the common factor Industrialization at time 1 (1960). Due to the repeated measurement design, the unique factors belonging to
and
for
are set to correlate. Additionally, the unique factors belonging to
and
for
are set to correlate. Indicators
and
serve as marker variables. The path diagram of the model is shown in .
Figure 3. Bollen (Citation1989).
Results of the estimation are presented in . The results in this example generally confirm the results from the previous example. In this case, the number of iterations and function evaluations are (It, Fe) (26, 759) for SNLLS and (It, Fe)
(230, 14742) for NLLS. shows the convergence profiles for the two implementations. The patterns in the figure resemble those in . Considering the estimation time, the mean time is 0.1257 sec. for SNLLS and 0.8263 sec. for NLLS. In this case, SNLLS proves to be faster by a factor of 0.8263/0.1257
6.5736.
Figure 4. Convergence profile, Bollen (Citation1989).
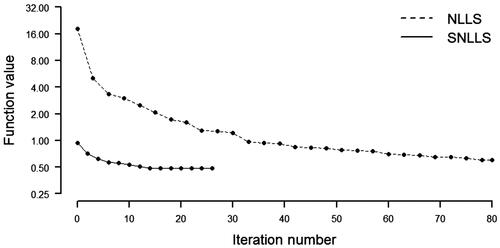
Table 2. Timing results, Bollen (Citation1989).
5. Concluding Remarks
In this study, we have presented an SNLLS implementation of the MD objective function for estimating SEMs. The outlined framework includes all necessary expressions for applying SNLLS, and represents a generalization of previously known results. Using examples from the SEM literature, we demonstrated that the computational load of applying SNLLS is considerably less than that of applying NLLS. Another benefit of SNLLS is that fewer starting values are required, which may mitigate potential problems due to the somewhat arbitrary choice of starting values for the covariance parameters.
The present work may have several interesting extensions. First, as shown by research, SNLLS may hold a potential for improving numerical performance in situations in which the estimation problem is ill-conditioned. Thus, an interesting case for future research would be to compare the numerical performance of SNLLS and NLLS under more challenging conditions in which the condition number of the observed covariance matrix is large. Second, the SNLLS implementation is not yet available for maximum likelihood (ML) estimation. Some initial work on this topic is underway. This work, combined with the previous point, may lead to an improved implementation of the ML estimator when applied to SEMs.
References
- Bollen, K. A. (1989). Structural equations with latent variables. Wiley.
- Dattner, I., Ship, H., & Voit, E. O. (2020). Separable nonlinear least-squares parameter estimation for complex dynamic systems. Complexity, 2020, 1–11. https://doi.org/10.1155/2020/6403641
- Geiser, C. (2012). Data analysis with Mplus. Guilford Press.
- Golub, G. H., & Pereyra, V. (1973). The differentiation of pseudo-inverses and nonlinear least squares problems whose variables separate. SIAM Journal on Numerical Analysis, 10, 413–432. https://doi.org/10.1137/0710036
- Golub, G. H., & Pereyra, V. (2003). Separable nonlinear least squares: The variable projection method and its applications. Inverse Problems, 19, R1–R26. https://doi.org/10.1088/0266-5611/19/2/201
- Hägglund, G. (1982). Factor analysis by instrumental variables methods. Psychometrika, 47, 209–222. https://doi.org/10.1007/BF02296276
- Kreiberg, D., Marcoulides, K., & Olsson, U. H. (2021). A faster procedure for estimating CFA models applying minimum distance estimators with a fixed weight matrix. Structural Equation Modeling: A Multidisciplinary Journal, 28, 725–739. https://doi.org/10.1080/10705511.2020.1835484
- Kreiberg, D., Söderström, T., & Yang-Wallentin, F. (2016). Errors-in-variables system identification using structural equation modeling. Automatica, 66, 218–230. https://doi.org/10.1016/j.automatica.2015.12.007
- Magnus, J. R., & Neudecker, H. (1999). Matrix differential calculus with applications in statistics and econometrics. John Wiley & Sons.
- MATLAB version R2020b. (2020). The MathWorks Inc.
- Mullen, K. M. (2008). Separable nonlinear models: Theory, implementation and applications in physics and chemistry [Ph.D. thesis]. Vrije Universiteit Amsterdam.
- R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org/
- Rosseel, Y. (2012). lavaan: An R package for structural equation modeling. Journal of Statistical Software, 48, 1–36. https://doi.org/10.18637/jss.v048.i02
- Sjöberg, J., & Viberg, M. (1997). Separable non-linear least-squares minimization-possible improvements for neural net fitting. In Neural networks for signal processing VII. Proceedings of the 1997 IEEE signal processing society workshop (pp. 345–354). IEEE.
- Söderström, T., & Mossberg, M. (2011). Accuracy analysis of a covariance matching approach for identifying errors-in-variables systems. Automatica, 47, 272–282. https://doi.org/10.1016/j.automatica.2010.10.046
- Söderström, T., Mossberg, M., & Hong, M. (2009). A covariance matching approach for identifying errors-in-variables systems. Automatica, 45, 2018–2031. https://doi.org/10.1016/j.automatica.2009.05.010
Appendices
A. Deriving L and K
Let be an arbitrary
random vector, and let
be the associated
covariance matrix. The purpose of the following presentation is to introduce an algebraic framework that facilitates eliminating the redundancy originating from the structure of
To do so, let
be a matrix such that
(A1)
(A1)
where
is a covariance vector containing the nonredundant elements of
and
is a matrix obtained by
(A2)
(A2)
In this expression,
is a selection matrix (i.e., a matrix composed of zeros and ones).
Below, we propose a rather general framework that applies to any structure characterizing Before presenting some examples on how to obtain
it is necessary to introduce some additional notation. Let
denote a
matrix (for
) with elements
(A3)
(A3)
Next, we demonstrate how to obtain
for two standard cases and one case specialized for the SNLLS implementation.
Case 1:
As a start, consider the case in which is symmetric and no other restrictions are placed on its elements. The covariance vector containing the
nonredundant elements of
is
(A4)
(A4)
Applying (A3), the matrix
is formed by horizontally concatenating
vectors using
(A5)
(A5)
Case 2:
Now, consider the case in which is diagonal. The covariance vector containing the
nonredundant elements of
is given by
(A6)
(A6)
The matrix
is now formed by horizontally concatenating
vectors
(A7)
(A7)
Before introducing the third and final case, it is necessary to expand the notation. Let
and
be respectively
and
random vectors, and let
be a
dimensional column vector obtained by stacking
and
in the following way
(A8)
(A8)
The associated p × p covariance matrix is given by
(A9)
(A9)
Case 3:
As in the first case, suppose that no other restrictions, apart from symmetry, are placed on the elements of Let the covariance vector containing the
nonredundant elements of
be given by
(A10)
(A10)
where
(A11)
(A11)
(A12)
(A12)
(A13)
(A13)
Construct a matrix by horizontally concatenating three matrices
(A14)
(A14)
where the submatrices are given by
(A15)
(A15)
(A16)
(A16)
(A17)
(A17)
The number of columns in (EquationA15
(A15)
(A15) ), (EquationA16
(A16)
(A16) ), and (EquationA17
(A17)
(A17) ) is
and
respectively.
The derivation below uses the following matrix identity
(B1)
(B1)
where
, and
are matrices of compatible sizes. In addition, we make use of the following relations
(B2)
(B2)
The model-implied covariance matrix is
(B3)
(B3)
Applying SNLLS, the key is to express the model-implied covariance vector using the form
(B4)
(B4)
To do so, it is necessary to vectorize the individual blocks of (B3). Starting with the block
we have
(B5)
(B5)
Using (EquationB1
(B1)
(B1) ) and (EquationB2
(B2)
(B2) ), it follows that
(B6)
(B6)
Next, we consider the block
Using the same procedure as before, we have
(B7)
(B7)
Finally, for the block
it follows that
(B8)
(B8)
Putting the pieces together, we obtain
(B9)
(B9)
