Abstract
A Bacillus strain having a certain extent of asymmetric resolution for (S)-ketoprofen was studied for its culture and conversion condition. The distribution and properties of lipases/esterases from its bacterial cell and the change of asymmetric resolution ability were also investigated. After purification by hydrophobic interaction chromatography and gel filtration, the lipase/esterase activity of active fraction was 61.7 times higher than the crude enzyme and the purity of 23 kDa protein increased more than 400 times. Hydrolytic activity of lipase/esterase using ketoprofen chloroethyl ester and pNPA as substrate remained essentially constant in the reaction system during the purification procedure. The above result clearly indicated that the application of hydrophobic interaction chromatography-gel filtration was fast, useful and effective when compared to the difficulties in purification of low content crude enzyme prepared directly from bacterial cells. Experimental results also indicated that there are several lipases/esterases in the bacillus cell selective to the optically active ketoprofen and the conversion result was derived from the comprehensive function of all these enzymes. This is a new way to understand the bacterial asymmetric resolution with high conversion rate and low optical purity. The ESI-QUAD-TOF mass spectrum was used to analyze 23 kDa protein. Protein identities were revealed by searching sequence databases (MSDB) with peptide sequence tags. The result showed that 23 kDa protein is a novel protein.
INTRODUCTION
In recent years, there has been a growing interest in the use of optically pure enantiomers for drugs, because they are more target-specific and have less side effects than racemic mixture [Citation[4]]. 2-Arylpropionic acids are an important class of non-steroid anti-inflammatory drugs. Ketoprofen is one of the most commonly prescribed members of this family. The (S)-enantiomer of Ketoprofen is 160–fold more active than its (R)-enantiomer for acesodyne and anti-inflammation.
In some cases, preparation of optically active enantiomers was accomplished by using microorganisms [Citation[7]] and enzymes. Hydrolytic enzymes such as esterase and lipase have been widely applied for the kinetic resolution of racemic acids and the use of biological systems for production of optically enriched compounds is becoming an alternative to the classical methodologies of chemical synthesis [Citation[8]]. Currently, the enantioselective resolution of 2-Arylpropionic acids for their optically active enantiomers by lipases from microorganisms has been the subject of intensive investigations.
The aim of the present study is to investigate the purification and properties of the lipases/esterases from a Bacillus strain screened for the asymmetric resolution of (S)-Ketoprofen. It is the foundation of consecutive construction of engineering bacteria for enantioselective resolution of (s)-ketoprofen. The study indicated that there are several lipases/esterases in the Bacillus strain with different selectivity to the optically pure ketoprofen. These results provided a better understanding to the comprehensive function of lipases/esterases in microorganisms for its asymmetric resolution.
MATERIALS AND METHODS
Materials
Strain NK13
In our previous work [Citation[9]], seventeen bacterial strains were screened from the soil in the vicinity of Naproxen–producing factory in China, from which 14 strains possessed lipase/esterase activity. One of the 14 strains was screened for a certain extent asymmetric resolutionof (S)-Ketoprofen and named as NK13.
Reagents
Ketoprofen chloroethyl ester was synthesized as described by Moreno [Citation[4]]. p-Nitrophenyl acetate (pNPA) was synthesized as described by Han [Citation[2]]. Phenyl Sepharose CL-4B and Sephacryl S-200 were purchased from Pharamacia. All other chemicals and solvents were of analytical grade.
Methods
Determination of Strain NK13
Strain NK13 was identified by Gram staining and spores staining. Its 16 S rRNA gene was also cloned and sequenced for comparison to those of other stains in GenBank.
Optimizing the Culture Condition of Strain NK13
Strain NK13 was grown at 37°C in a solid medium that contained (per liter) 5 g of peptone, 5 g of NaCl, 10 ml of Tween-20, and 15 g of agar (pH 7.0), then grown aerobically at 30°C in a liquid medium that contained (per liter) 12 g of glucose, 5 g of yeast extract paste, 1 g of peptone, 5 g of NaCl, and 5 ml of Tween-20 (pH 7.0). Changing the pH and substituting Tween-80 for Tween-20 in a liquid medium optimized the cultural conditions of the bacterial. Finally, the bacterial growth curves were established.
Assays for Lipase/esterase Activity
Photometric Assay
Photometric assay was performed as described by U. Bornscheuer [Citation[1]]. p-nitrophenyl acetate (pNPA) was used for a quick analysis of crude lipase/esterase and chromatographic fractions. Solution A contained 90 mg p-nitrophenyl acetate (pNPA) and was dissolved in 30 ml 2-propanol with a sonicator for 6 min at room temperature. Solution B contained 0.4% Triton-X-100 and 0.1% gum arabic. Both solutions are stable for about 2 weeks in a refrigerator. The reaction mixture consisted of 1 part solution A and 9 parts solution B, which was freshly prepared before each assay. A 100 µl volume of an appropriate dilution of the enzyme solution was added to 900 µl of the reaction mixture. Finally, the lipase reactions were analyzed at 410 nm and 30°C with an Ultrospec-K photometer. One unit of lipase/esterase activity was defined as the amount of enzyme that liberated 1 µmol p-nitrophenol per min from pNPA.
Protein Determination
Analysis was performed as described by U. Bornscheuer et al. [Citation[1]] with 0.1% Coomassie Brilliant Blue in 1% acetic acid, 40% methanol.
Lipase Hydrolysis Reaction System
The reaction system contained 1 ml lipase (dissolved by 25 mmol/l PBS, pH 7.0) and 5 µl 20% ketoprofen chloroethyl ester (dissolved by ethyl acetate) was cultured in a shaker of 140 rpm at 30°C. After the reaction, the pH value was adjusted below 3 with 4 M HCl and the desired product was extracted with 1.2 ml ethyl acetate.
Isolation and Analysis of Ketoprofen
The extract obtained from lipase/esterase reaction system was assayed by thin-layer chromatography (TLC) and its conversion yield was obtained by IOD analysis to TLC panel. The product band was then separated from the panel, extracted with ethyl acetate. Optical purity (ee%) of product (S)-ketoproxen was assayed by HPLC (Water 600E). Chiral column, CHIROBIOTICV; Mobile phase, tetrahydrofuran: citric acid/sodium citrate (pH 6.3, 0.05 mol/L) = 10:90 (V/V); Effluence rate:1 ml/min.
SDS-PAGE Assay
Discontinuous SDS-PAGE and determination of the enzyme were performed with low molecular weight standard mixtures as described by J. Sambrook et al. [Citation[5]]. After electrophoresis gel were stained with AgNO3 by the method of Heukeshoven [Citation[3]] and scanned by densitometry.
Preparation of Crude Lipase/Esterase
Precipitate of bacterial cells was harvested from 1000 ml cultures by centrifugation in a high-speed centrifuge at 5000 rpm for 10 min. After weighing the precipitate, it was dissolved by 25 mM PBS/1 mM EDTA (2 ml buffer per g bacterial cells, pH 7.0) and stored at −20°C. The bacterial cells were thawed at 20°C immediately and lysozyme was added (10 mg lysozyme per g bacterial cells), then placed in a water bath at 37°C for 20 min. The cells were levigated thoroughly by quartz sand at 0°C, and the supernatant (crude lipase/esterase) was obtained by centrifugation in a high-speed centrifuge at 12000 rpm for 20 min at 4°C.
Purification of Lipase/Esterase by Hydrophobic Interaction Chromatography
Crude lipase/esterase was loaded on a Phenyl Sephrose CL-4B column (2.0 cm × 25.0 cm), equilibrated with 50 mM piperazine buffer (pH 7.0). Unbound compounds were washed out with 150 ml piperazine buffer and, finally, the lipases/esterases were eluted with a linear gradient of isopropanol at a flow rate of 1 mL/min. Linear gradients of eluted isopropanol fractions were obtained (4 ml per tube) and assayed at 280 nm.
Further Purification of Lipase/Esterase by Gel Filtration
The active fractions from hydrophobic interaction chromatography were pooled and dialyzed against deionized water for 20 h. After concentrated by freeze dryer, the dialysates were prepared to contain 3 mg/mL in 25 mM PBS (pH 7.0) and then applied to a Sephacryl S-200 column (1.6 cm × 70 cm) equilibrated with 25 mM PBS (pH7.0). The enzymes were eluted by adding 25 mM PBS (pH 7.0) at a flow rate of 0.2 mL/minand 40 ml to 100 ml was obtained (2 ml per tube).
ESI-QUAD-TOF MS
Protein bands excised from SDS-PAGE gels were destained, reduced, alkylated, and then in-gel digested with trypsin as described by Shevchenko et al. [Citation[6]]. The extracted peptides were analyzed on a quadrupole/time-of-flight mass spectrometer (QSTAR, PE Sciex) equipped with a nanoelectrospray ion source. Protein identities were revealed by searching sequence databases with peptide sequence tags generated by tandem mass spectrometry.
RESULTS AND DISCUSSIONS
Determination of Strain NK13
NK13 strain was characterized to be a Gram-positive Bacilli with spores. It is active and aerobic, 2–3 µm wide, 4–6 µm long. 16 rRA determination (AY654879) further revealed its high homologity to Bacillus strains: in total 32 strains, which had a homologity >99%, 30 strains are the members of Bacillus strains, and the other two were unidentified Gram-positive bacterial. The above results indicated that NK13 belongs to Bacillus strain.
Optimizing the Culture Condition of Strain NK13 and Locating the Lipases/Esterase
The culture conditions of NK13 strain were studied. Both in the presence of Tween-20 and Tween-80, the cell growth increased with increase in pH value, though the growth were little different at a pH range from 7.5–9, experimental results signified that the optimal cell culture pH was 8.5.
The growth curve of NK13 strain at 30°C, pH = 7 and Tween-80, reached logarithm growth at 5–7 h of culture. Compared to the logarithm growth using Tween-20 (growth time 8–10 h), the growth time with Tween-80 was shortened by 3 h and the strain was more bloomed.
After centrifugation of cultures, lipase activity was assayed respectively in the supernatant and in the precipitate (bacterial cells) with 25 mM PBS (pH 7.0) in different reaction times. Experimental results showed that the lipases were in the bacterial cell. Consecutive hydrolysis conversion reactions further proved their location.
Purification and Analysis of Lipase/Esterase from Strain NK13
Piperazine-isopropanol was used as a continuous gradient eluent, the lipases/esterases were eluted concentratively in fraction tubes 16–18 which corresponded to a small 280 nm protein peak. The concentration of lipase in isopropanol was 19.3%–26.0%.
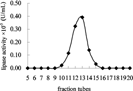
Fraction tubes of 16–18 from hydrophobic interaction chromatography were pooled as active fraction and concentrated, then applied to a Sephacryl S-200 column. showed that the lipases/esterases were further purified and as a peak eluted concentratively in fraction tubes 10–15. The result by 280 nm protein scanner indicated that this lipase activity peak corresponded to the latter part of a small 280 nm protein peak.
Figure 1 Lipase/esterase activity assay to eluted faction of gel filtration by Sephacryl S-200 column.
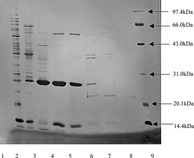
showed the SDS-PAGE assay result. It indicated that after purification by hydrophobic interaction chromatography the impure protein decreased greatly compared to crude lipase and further purification by gel filtration gave a better result. There were few proteins in the active fraction tubes 10–15. Especially, there was only 23 kDa protein in tube 15. It was proved that this is a lipase/esterase with enantioselective resolution of (s)-ketoprofen by consecutive conversion experiments.
Figure 2 SDS-PAGE assay of lipases/esterases from NK13 strain. SDS-PAGE lanes: 1, crude enzymes; 2, active fraction from hydrophobic interaction chromatography; 3,4,5,6,7,8 correspond to fraction tube 10,11,12,13,14,15 from gel filtration, respectively; 9, molecular weight standards.
showed the differences of specific lipase/esterase activity among the crude lipases, active fraction from HIC and active fraction tubes 10–15 from gel filtration. After a single HIC, the specific lipase activity was 16.7-fold higher than the crude one. Further purification by gel filtration increased 3.70-fold on the basis of purification by HIC.
Table 1. Purification result of lipases from NK13 strain
revealed that the purity of 23 kDa protein had an increase of 17.3-fold by HIC and a single band in tube 15 by SDS-PAGE assay.
Table 2. Purity of 23 kDa protein
The above result clearly indicated that the application of hydrophobic interaction chromatography-gel filtration was fast, useful and effective when compared to the difficulties in purification of low content crude lipase prepared directly from bacterial cells.
and showed the conversion rate and optical purity of ketoprofen from crude enzyme, active fraction from HIC and active fraction tubes from gel filtration. After the Bacillus bacterial cells were processed by lysozyme and quartz sand, the crude enzyme reacted 24, 48, 72 hours, respectively, in PBS conversion system. Experimental results showed that the conversion rate had a little difference in the above three periods of reaction time but the 24 hours' optical purity of (S)-ketoprofen was better, which reached 6.18%. The conversion rate of active fraction by HIC also showed little differences in the three periods of time and the 24 hours' optical purity of (S)-ketoprofen was 18.9%, which was still the best one. These results dropped a hint that there was more than one lipase/esterase with asymmetric resolution capability.
Table 3. Comparison of conversion rate and optical purity of ketoprofen by crude lipases and active fraction from HIC
Table 4. 72 h conversion rate of ketoprofen by crude lipases, active fraction from HIC and active fraction tubes from gel filtration
Although using different kinds of proteins in the tubes containing active fractions, (S)-ketoprofen could be obtained by asymmetric resolution after gel filtration. Experimental results showed there was only 23 kDa protein in tube 15 and its conversion rate was 52%. As to other active fraction tubes, their ee (%) of (S)-ketoprofen were different and did not correspond to their lipase activity. also revealed that the location and content of 55 kDa, 39 kDa, 38 kDa and 33 kDa proteins were different to each active fraction tubes from gel filtration. These results indicated that there were several lipases selective to the optically active ketoprofen and the conversion result was derived from the comprehensive function of all these lipases/esterases. In conclusion, 55 kDa protein is a lipase/esterase for enantioselective resolution of (S)-ketoprofen and 39 kDa, 38 kDa, 33 kDa proteins are lipases/esterases for enantioselective resolution of (R)-ketoprofen. The above results provided a new way to understand the bacterial asymmetric resolution with high conversion rate and low optical purity.
Analyzing 23 kDa Protein by ESI-QUAD-TOF MS
The ESI-QUAD-TOF mass spectrum was used to analyze 23 kDa lipase/esterase. Protein identities were revealed by searching sequence database (MSDB) with peptide sequence tags. The result showed that 23 kDa lipase/esterase is a noval protein. Searches in other protein databases such as Swiss-Prot/TrEMBL and PROSITE also revealed that there was no identical lipase/esterase sequence and conserved sequence of Bacillus lipase (Gly/Ala-X1-Ser-X2-Gly) in peptide sequence tags. Considering that the peptide sequence provided by mass spectrum was only part of the complete sequence, the conserved sequence was estimated to be a part of the residue and the above Ms results of 23 kDa protein in Bacillus NK13 built a solid foundation to consecutive construction of engineering bacteria for enantioselective resolution of (S)-ketoprofen.
REFERENCES
- Bornscheuer, U., Reif, O.W., Lausch, R., Freitag, R., Scheper, T., Kolisis, F.N., Menge, U. (1994). Lipase of Pseudomonas cepacia for biotechnological purposes: purification, crystallization and characterization. Biochim. Biophys. Acta. 1201(1): 55–60. [PUBMED], [INFOTRIEVE], [CSA]
- Han, G.D., Fan, R.L., Li, S.W. (1978). Chemical Handbook of Organic Synthesis, 2nd ed., Petroleum Chemical Industry Publication: Beijing.
- Heukeshoven, J., Dernick, R. (1985). Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6: 103–112. [CSA], [CROSSREF]
- Moreno, J.M., Sinisterra, J.V. (1995). A systematic analysis of the variables that control a highly stereoselective resolution of racemic non-steroidal antiinflammatory drugs using immobilized lipase from Candida cylindracea. J. Mol. Catal. A: Chemical 98(3): 171–184. [CSA], [CROSSREF]
- Sambrook, J., Fritsch, E.F., Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press: New York, pp. 636–643.
- Shevchenko, A., Wilm, M., Vorm, O., Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68(5): 850–858. [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Yamamoto, K., Oishi, K., Fujimatsu, I., Komatsu, K.I. (1991). Production of R-(-)-mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl. Environ. Microbiol. 57(10): 3028–3032. [PUBMED], [INFOTRIEVE], [CSA]
- Zhang, J.H., Hou, Z.B., Yao, C.Y., Yu, Y.T. (2002). Purification and properties of lipase from a Bacillus strain for catalytic resolution of (R)-Naproxen. J. Mol. Catal. B: Enzymatic 18(4–6): 205–210. [CSA], [CROSSREF]
- Zhang, J.H., Yao, C.Y., Yu, Y.T., He, Z.H., Yang, X.Q., Yang, W.B. (1999). Preliminary investigation of enantioselective hydrolysis of chiral ester from microorganism. Acta Scientiarum Naturalium Universitatis Nankaiensis 32(4): 39–41. [CSA]