Abstract
The karyotype of the giant anteater Myrmecophaga tridactyla was studied throughout the species’ Argentine range. Images of the chromosome complement clearly revealed the karyotype and identified previously misinterpreted chromosomes. The peripheral blood lymphocytes of 26 adult animals (11 males and 15 females) were cultured and used to obtain mitotic metaphases. These preparations were subjected to conventional staining, G- and C-banding, and Fluorescent in situ Hibridization (FISH). Spermatocyte pachytene microspreads for one adult were examined for synaptonemal complexes. The karyotype (2n = 60 XX/XY; Fundamental Number (FN) = 110 without XY) showed an autosomal complement of six metacentric, 18 submetacentric, and six acrocentric chromosome pairs. The X chromosome (4.67 ± 0.29% of the haploid set) was shown to be large and submetacentric, similar in size to autosomes 3–5. The Y chromosome was submetacentric (2.60 ± 1.08% of the haploid set). It is, however, larger than the Y chromosome of the closely related armadillos. Pairs 1, 3, 4, 5, 6, 9, 11, 14, 15, 23, 25 and 29 showed small masses of heterochromatin in the pericentromeric region. These masses were slightly larger in chromosome pairs 8, 10 and 18. Pairs 2, 7, 16, 17, 19, 20, 21, 22, 24, 26, 27 and 28 were entirely C-negative. Analysis of the telomeric sequences by FISH involving a Cy3-conjugated peptide nucleic acid pantelomeric probe revealed no signals from the interstitial regions. Ag-NORs were located on chromosomes 5, 10, 11, 21 and 23. The spermatocyte pachytene microspreads confirmed the morphology and size of both sex chromosomes. The present results, obtained via the analysis of a large number of specimens, provide an in-depth characterization of the chromosomal complement of this species.
Introduction
The extant members of Xenarthra – eminently Central and South American placental mammals – form three phylogenetic groups: the arboreal sloths (Folivora: Megalonychidae and Bradypodidae), the shelled armadillos (Cingulata: Dasypodidae) and the toothless anteaters (Vermilingua: Myrmecophagidae and Cyclopedidae) (Gardner Citation2008). Chromosomal polymorphisms, pairs of heteromorphic autosomes of different lengths (Benirschke & Wurster Citation1969; Jorge et al. Citation1978; Rossi et al. Citation2014), and unusual sex chromosome systems (Corin-Frederic Citation1969; Jorge et al. Citation1985) have been described in some species of Xenarthra. A number of cytogenetic studies have been performed but none have involved large numbers of specimens from different locations.
The family Myrmecophagidae contains two genera: Myrmecophaga and Tamandua. Myrmecophaga comprises only one species, the giant anteater Myrmecophaga trydactyla, which has, according to preliminary reports, 60 chromosomes, some of which are acrocentric (Hsu Citation1965; Pereira et al. Citation2004). Tamandua has two species of vested anteater, Tamandua tetradactyla and Tamandua mexicana, neither of which have acrocentric chromosomes. Two chromosome numbers have been reported for T. tetradactyla (Pereira et al. Citation2004). Diploid numbers for the family Myrmecophagidae range from 2n = 54 to 2n = 60. Reciprocal translocations and reversible fusions/fissions may explain these differences (Jorge et al. Citation1985).
Myrmecophaga tridactyla – the giant anteater – is recognized as vulnerable by the ISCN (Shaffer et al. Citation2009). Although the karyotype of this species has been described in two studies, both involved only conventional staining techniques and just a few individuals of unknown provenance living in captivity (Hsu Citation1965; Pereira et al. Citation2004). For example, no G-banding patterns were reported that might allow the sex chromosomes to be identified. Thus, if the karyotype of this species is to be reliably understood, studies involving cytogenetic and resolution banding techniques are required. This study reports the complete cytogenetic characterization of M. tridactyla. Mitotic spreads were examined by G- and C-banding, Nucleolus Organizer Región (NOR) staining and Fluorescent in situ Hibridization (FISH), and meiotic spreads via synaptonemal complex (SC) analysis.
Materials and methods
Sampled specimens
The animals studied were adult specimens (11 males and 15 females) of M. tridactyla housed at various zoos, breeding centres and wildlife reintroduction facilities in different parts of Argentina (). After immobilization using Telazol (4 mg/kg) with a Telinject (remote injection) system, peripheral blood samples were collected with disposable heparinized syringes. All handling was performed according to the Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research (Sikes et al. Citation2011).
Table I. List of studied individuals with their respective sex and localities of provenance.
Mitotic preparations
Lymphocytes were cultured for 72 h at 34°C in RPMI 1640 Medium (Roswell Park Memorial Institute, Gibco) according to Moorhead et al. (Citation1960). Metaphase chromosome spreads were prepared following the usual air-drying methods, and then stained with carbol fuchsine (Carr & Walker Citation1961). G-banding was performed following the method of Wang and Fedoroff (Citation1972), which involves trypsin treatment and subsequent staining with 5% Giemsa solution at pH 6.8. C-banding was performed according to Sumner (Citation1972). The slides were incubated with a supersaturated solution of barium hydroxide (BaOH) at 50°C, and then further incubated for 1 h in 2XSSC solution at 60°C. They were then stained with 1.5% Giemsa solution at pH 6.8 for 15 min (Sumner Citation1972). G-banding was carried out in not less than five metaphases for each specimen. At least 10 G- and 10 C-banded metaphases were selected to be used in the production of ideograms.
Silver nitrate staining of mitotic and meiotic material was performed according to Sciurano et al. (Citation2006). All preparations were photographed using a Leitz DMRB microscope (Leica Microsystems, Wetzlar, Germany) and a Leica DFC 300 FX digital camera (Leica Microsystems, Cambridge, UK).
Ideograms were prepared for all chromosomes. The morphology of the individual chromosomes of three cells was characterized and karyotypes assembled in the conventional manner (Shaffer et al. Citation2009).
Fluorescence in situ hybridization
The telomeric distribution of two complementary oligonucleotides (Telo1, TTAGGG7 Oligo number 203006A623H01 ½, and Telo2, GGGTTA7 Oligo number 20306A623H02 2/2) was analysed by FISH in metaphase spreads, using a Cy3-conjugated peptide nucleic acid (PNA) pantelomeric probe (Biofab Research, Roma, Italy). FISH of the repetitive sequences was performed following standard procedures (Lichter et al. Citation1992). The slides were then conventionally stained with propidium iodide and mounted in Vectashield medium (Vector Laboratories, Peterborough, UK). DAPI (4’,6-diamidino-2-phenylindole) counterstaining facilitated the identification of homologues. Signals were observed at 1000 × using a Leica DM epifluorescence microscope (Leica Microsystems, Wetzlar, Germany) equipped with an HBO 50 mercury lamp and filters for DAPI and Cy3 (Chroma Technology, Bellows Falls, USA). A Leica DFC 300 FX digital camera (Leica Microsystems) was used for all photography. Images were processed using Adobe Photoshop CS software (Adobe Systems Inc.).
Meiotic bivalent and sex chromosome analysis
Testicular material was obtained from an animal (specimen number 25) that died of natural causes. Spermatocyte microspreads for SC analysis were prepared as described by Sciurano et al. (Citation2012). Some slides were stained with 4% phosphotungstic acid in ethanol or silver nitrate (Howell & Black Citation1980), while others were kept at −70°C until examination by immunofluorescence microscopy. The immunolocalization of meiotic proteins was performed according to Sciurano et al. (Citation2012). The following primary antibodies were used: rabbit anti-SMC3 (1:500) (Merck Millipore, Billerica, MA, USA) to recognize components of the cohesion complex and thus identify bivalents; and human CREST serum that binds to kinetochores (1:10) (Laboratorios IFI, Buenos Aires, Argentina). Slides were examined using a LEICA DM epifluorescence microscope (Leica Microsystems) and photographed with a Leica DFC 300 FX digital camera (Leica Microsystems). Separate images were superimposed using Adobe Photoshop CS software (Adobe Systems Inc.).
Results
Mitotic studies
All individuals showed a chromosome number of 60 (58 + XX/XY; female/male), with six metacentric and 18 submetacentric chromosome pairs, all clearly two-armed, plus six acrocentric pairs (). The fundamental number (FN) was 110. The only difference in the karyotypes was the morphology of chromosome pairs 2 and 3, which were heteromorphic in 65% of individuals. This polymorphism, involving the absence of small arms, is probably the result of pericentromeric inversion.
Figure 1. (a), Karyotype of a male Myrmecophaga tridactyla from Alberdi, Province of Santiago del Estero. (b), G-banded karyotype of M. tridactyla from Copo, Province of Santiago del Estero. (c), Ideogram of a male M. tridactyla based on the G-banded karyotype Scale bars: 10 µm.
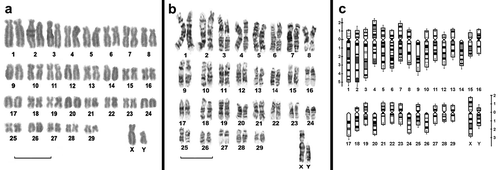
The X chromosome (4.67 ± 0.29% of the haploid set) was large and submetacentric, and similar in size to autosomes 3–5. The Y chromosome was submetacentric (2.60 ± 1.08% of the haploid set) and it is between medium and smaller chromosomes (). G-banding accurately identified all the autosomal pairs and the X and Y chromosomes. The homology between the two elements of each pair and the X and Y chromosome was checked by G-banding comparisons (). The ideogram () is a composite representation of all the G-bands seen in 10 well-banded karyotypes (it indicates the average location of each of the bands detected).
The C-banding pattern in pairs 1, 3, 4, 5, 6, 9, 11, 14, 15, 23, 25 and 29 showed small masses of heterochromatin in the pericentromeric region; these were slightly larger in pairs 8, 10 and 18. Other pairs were entirely C-negative (pairs 2, 7, 16, 17, 19, 20, 21, 22, 24, 26, 27 and 28). C-banding analysis revealed no pericentromeric constitutive heterochromatin in the X and Y chromosomes ().
Figure 2. (a), C-banded karyotype of a male Myrmecophaga tridactyla from the Province of Santiago del Estero, showing centromeric C+ heterochromatin. (b), NOR-banded karyotype of a male M. tridactyla from the Province of Salta. Identification of the NORs (Arrowhead). (c), SC karyotype of a male M. tridactyla from Tostado, Province of Santa Fe. Each bivalent is represented by 1 SC immunolabeled with anti-SMC3 (red), anti-kinetochore (yellow). (d), Metaphase plate of a male M. tridactyla from the Province of Salta, showing the location of the telomeric probes. The chromosomes were counterstained with DAPI. Scale bars: 10 µm.
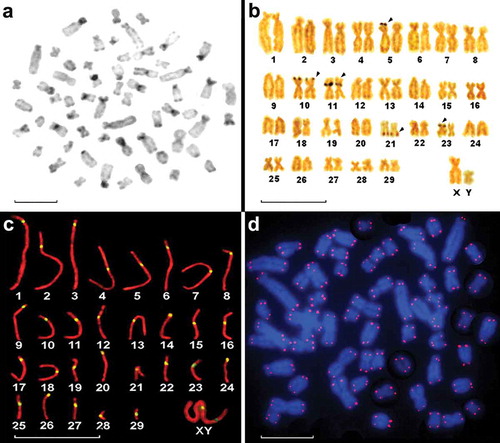
Five nucleolar organizing regions (Ag-NORs), one on each of chromosome pairs 5, 10, 11, 21 and 23, were observed in most cells (). The SC karyotype showed 29 autosomal SCs, 24 autosomal SCs with meta or submetacentric components and five autosomal SCs with acrocentric elements. The location of the centromeres (kinetochores) was observed and verified by inmunolocalization (). The spermatocyte pachytene microspreads corroborated the chromosomal morphology and FN () and clearly showed the sex chromosome kinetochores. Hybridization of the probe with the telomeric sequences revealed no regions with interstitial signals ().
Discussion
The present work ascertained the karyotype of M. tridactyla using classical and molecular cytogenetic techniques. All chromosomes were identified, including some that had previously been misinterpreted.
All the animals examined had a chromosome complement of 2n = 60, confirming the results of other authors (Hsu Citation1965; Pereira et al. Citation2004). Pereira et al. (Citation2004) reported the giant anteater to have five pairs of small- and medium-sized acrocentric chromosomes. In the present work, however, six pairs of acrocentric chromosomes were seen; one of the medium-sized submetacentric pairs described by the latter authors was in fact a medium-sized acrocentric pair.
The sex chromosomes showed morphologies different to those described by Pereira et al. (Citation2004) for giant anteaters from Brazil. In the present work, the X chromosome was large and submetacentric, and the Y chromosome was also submetacentric. It was larger than that of the closely related armadillos; in armadillos, the Y chromosome is the smallest of the entire complement (Sciurano et al. Citation2012; Rossi et al. Citation2014). Pereira et al. (Citation2004) reported the Y chromosome to be small and acrocentric. G- and C- bandings confirmed the presence of chromosome designations and identified those erroneously paired by these authors.
The differences in the distribution of the C-positive regions in Myrmecophagidae species reflect the variation in the content, location and distribution of the constitutive heterochromatin in this family and within Xenarthra (Dobigny et al. Citation2005; Liu et al. Citation2011; Rossi et al. Citation2014). M. tridactyla is characterized by the presence of small masses of heterochromatin in most of its chromosomes, whereas T. tetradactyla has five pairs of chromosomes with large centromeric heterochromatin regions (Dobigny et al. Citation2005). Interestingly, no telomeric signals were found in M. tridactyla, whereas T. tetradactyla has five pairs of chromosomes that show interstitial telomeric signals (unpublished results). It is well known that clusters of different sequences of repetitive DNA, including telomeric repeats in subtelomeric and interstitial positions, are useful for characterizing the breakpoints of recurrent chromosomal rearrangements (Azzalin et al. Citation2001; Nergadze et al. Citation2004). The C-banding pattern and telomeric sequence of M. tridactyla may clarify the chromosomal mechanisms underlying the karyotype evolution of the family Myrmecophagidae. The difference in chromosome number between M. tridactyla and T. tetradactyla species might be explained by Robertsonian translocation.
Species with more primitive karyotypes show the smallest number of NORs (Chiarelli & Capanna Citation1973). It is well documented that different groups of species have acquired more NORs over time (Hall & Parker Citation1995; Hirai et al. Citation1996). Among the Xenarthrans, Dasypus hybridus (2n = 64, FN = 79) (Saez et al. Citation1964) has one NOR (Sciurano et al. Citation2006), Chaetophractus villosus has three (Sciurano et al. Citation2006; Rossi et al. Citation2014) and M. tridactyla has five (as revealed by the Ag-NOR technique).
The SCs in spermatocytes examined during early prophase showed the X and Y chromosomes of M. tridactyla to be submetacentric, with a clearly defined short arm. This was not reported by previous authors (Hsu Citation1965; Pereira et al. Citation2004).
The present results update the available information on the karyotype of the giant anteater. Modern cytogenetic techniques may be able to show how changes in the chromosomes affected the final complement of this species and the family Myrmecophagidae.
Acknowledgements
M.S. Merani was supported by grant PICT 1198 from the Agencia Nacional de Promoción Científica y Tecnológica and grant PIP 0204 from the Consejo Nacional de Investigaciones Científicas y Técnicas.
References
- Azzalin CM, Nergadze SG, Giulotto E. 2001. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 110:75–82. doi:10.1007/s004120100135.
- Benirschke K, Wurster DH. 1969. The chromosomes of the giant armadillo, Priodontes giganteus Geoffroy. Acta Zoologica et Pathologica Antverpiensia 49:125–130.
- Carr DH, Walker JE. 1961. Carbol fuchsin as a stain for human chromosomes. Stain Technology 36:233–236.
- Chiarelli AB, Capanna E. 1973. Cytotaxonomy and vertebrate evolution. New York: Academic Press.
- Corin-Frederic J. 1969. Les formules gonosomiques dites aberrantes chez les Mammifères Euthèriens. Chromosoma 27:268–287. doi:10.1007/BF00326165.
- Dobigny G, Yang F, O’Brien PCM, Volobouev V, Kovács A, Pieczarka JC, Ferguson-Smith MA, Robinson TJ. 2005. Low rate of genomic repatterning in Xenarthra inferred from chromosome painting data. Chromosome Research 13:651–663. doi:10.1007/s10577-005-1002-9.
- Gardner AL. 2008. Magnaorder Xenarthra. In: Gardner AL, editor. Mammals of South America, Volume 1: Marsupials, xenarthrans, shrews, and bats. Chicago: The University of Chicago Press. pp. 127–176.
- Hall KJ, Parker JS. 1995. Stable chromosome fission associated with rDNA mobility. Chromosome Research 3:417–422. doi:10.1007/BF00713891.
- Hirai H, Yamamoto MT, Taylo RW, Imai HT. 1996. Genomic dispersion of 28S rDNA during karyotypic evolution in the ant genus Myrmecia (Formicidae). Chromosoma 105:190–196. doi:10.1007/BF02509500.
- Howell WM, Black DA. 1980. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36:1014–1015. doi:10.1007/BF01953855.
- Hsu TC. 1965. Chromosomes of two species of anteaters. Mammalian Chromosomes Newsletter 15:108–109.
- Jorge W, Meritt DA Jr, Benirschke K. 1978. Chromosome studies in Edentata. Cytobios 18:157–172.
- Jorge W, Orsi-Souza AT, Best RC. 1985. The somatic chromosomes of Xenarthra. In: Montgomery GG, editor. The evolution and ecology of armadillos, sloths and vermilinguas. Washington and London: Smithsonian Institution Press. pp. 121–129.
- Lichter P, Boyle A, Wienberg J, Arnold N, Popp S, Cremer T, Ward DC. 1992. Nucleic acid hybridization. General aspects. In: Roche Diagnostics GmbH, editor. DIG Application Manual for Nonradioactive In Situ Hybridization (Application manual). Mannheim: Roche Diagnostics GmbH. pp. 28–33.
- Liu Y, Ye J, Fu B, Ng BL, Wang J, Su W, Yang F, Nie W. 2011. Molecular cytogenetic characterization of the Genome organization of the 6-Banded armadillo (Euphractus sexcinctus). Cytogenetic and Genome Research 132:31–40. doi:10.1159/000318706.
- Moorhead PS, Nowell PC, Mellman WJ, Battips DM, Hungerford DA. 1960. Chromosome preparations of leukocytes cultured from human peripheral blood. Experimental Cell Research 20:613–616. doi:10.1016/0014-4827(60)90138-5.
- Nergadze SG, Rocchi M, Azzalin CM, Mondello C, Giulotto E. 2004. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Research 14:1704–1710. doi:10.1101/gr.2778904.
- Pereira HRJ Jr, Jorge W, da Costa MELT. 2004. Chromosome study of anteaters (Myrmecophagideae, Xenarthra): A preliminary report. Genetics and Molecular Biology 27:391–394. doi:10.1590/S1415-47572004000300014.
- Rossi LF, Luaces JP, Alonso FM, Merani MS. 2014. Karyotype and chromosome variability in the Armadillo Chaetophractus villosus in Argentina. Cytogenetic and Genome Research 142:101–106. doi:10.1159/000357219.
- Saez FA, Drets ME, Brum N. 1964. The chromosomes of the mulita (Dasypus hybridus): A mammalian edentate of South America. In: Pavan C, editor. Mammalian cytogenetics and related problems in radiobiology. New York: Pergamon Press. pp. 163–170.
- Sciurano RB, Merani MS, Bustos J, Solari AJ. 2006. Synaptonemal complexes and XY behavior in two species of Argentinian armadillos: Chaetophractus villosus and Dasypus hybridus (Xenarthra, Dasypodidae). Biocell 30:57–66.
- Sciurano RB, Rahn MI, Rossi LF, Luaces JP, Merani MS, Solari AJ. 2012. Synapsis, recombination, and chromatin remodeling in the XY body of armadillos. Chromosome Research 20:293–302. doi:10.1007/s10577-012-9273-4.
- Shaffer L, Slovak ML, Campbell LJ, editors. 2009. ISCN. An international system for human cytogenetic nomenclature. Basel: S Karger.
- Sikes RS, Gannon WL, and the Animal Care and Use Committee of the American Society of Mammalogists. 2011. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy 92:235–253. doi:10.1644/10-MAMM-F-355.1.
- Sumner AT. 1972. A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75:304–306. doi:10.1016/0014-4827(72)90558-7.
- Wang HC, Fedoroff S. 1972. Banding in human chromosomes treated with trypsin. Nature: New Biology 235:52–54.
