Abstract
The filamentous Ascomycota Colletotrichum gloeosporioides sensu lato is a fungus that has been reported worldwide as a causal agent of anthracnose disease in avocado and other crops. In Mexico, this species affects fruits from an early stage of development in the orchard until the post-harvest stage. Although fungicides are continuously applied to control Colletotrichum species, pericarp cankers and soft rot mesocarp in fruits are still frequently observed. Considering the lack of a precise description of the causative agent, the aim of the current study was to determine the pathogens involved in this symptomatology. Twenty-four isolates were consistently obtained from the pericarp of avocado fruits cv. “Hass” collected in the central avocado-producing area of Mexico. Morphological features such as colony growth, conidia size, and mycelial appressorium were assessed. Bayesian multilocus phylogenetic analyses were performed using amplified sequences of the internal transcribed spacer region of the nuclear ribosomal DNA; actin, chitin synthase, glyceraldehyde-3-phosphate dehydrogenase partial genes; and APn2-Mat1-2 intergenic spacer and mating type Mat1-2 partial gene from the nine selected isolates. In addition, fruits were inoculated with a conidial suspension and reproducible symptoms confirmed the presence of Colletotrichum fructicola in this area. This pathogenic species can now be added to those previously reported in the country, such as C. acutatum, C. boninense, C. godetiae, C. gloeosporioides, and C. karstii. Disease management programs to reduce the incidence of anthracnose should include C. fructicola to determine its response to fungicides that are routinely applied, considering that the appearance of new species is affecting the commercial quality of the fruits and shifting the original population structure.
1. Introduction
Anthracnose is a disease caused by the Colletotrichum species complex, an intracellular filamentous Ascomycota that is characterized by the formation of sunken spots and canker in the stems, twigs, branches, and fruits [Citation1]. This pathogen affects many hosts worldwide such as avocado (Persea americana), annona (Annona cherimola), coffee (Coffea arabica), apple (Malus domestica), peach (Prunus persica), mango (Mangifera indica), and, more recently, tea oil (Camellia oleifera), among others [Citation2–6].
Colletotrichum gloeosporioides has been extensively studied, and a species complex comprising 22 species along with one subspecies using multilocus phylogenetic analysis was recently described [Citation7], and shown to be formed by 42 accepted species [Citation8–10]. This species complex has been reported as the causal agent of anthracnose in all avocado-producing countries, and represents the most serious post-harvest disease in high rainfall growing regions [Citation11]. Furthermore, avocado anthracnose has been reported to be caused by C. gloeosporioides in South Africa [Citation12] and Israel [Citation13], C. fioriniae in Australia [Citation14], C. aenigma in Israel, C. alienum in Australia and New Zealand, C. fructicola in Australia [Citation7], C. kahawae subsp. ciggaro in New Zealand, C. queenslandicum in Australia, and C. siamense in Australia and South Africa [Citation7].
In Mexico, C. gloeosporioides, C. boninense, C. acutatum [Citation15], C. godetiae [Citation16], and C. karstii [Citation17] have been isolated from avocado exocarp tissue with anthracnose symptoms. Nevertheless, there is no mention of C. gloeosporioides sensu stricto causing anthracnose in avocado in Mexico, which may be because previous studies only assessed C. gloeosporioides using a single locus based on the internal transcribed spacer (ITS) region of rDNA, with no comparison to newly redefined species belonging to C. gloeosporioides sensu lato. Indeed, the ITS region was proposed as a fungal barcode marker [Citation18] and has been used for Colletotrichum identification in several studies [Citation15,Citation19–21].
Nevertheless, multilocus phylogenetic analyses have clarified that the species included in the C. gloeosporioides complex are plant pathogenic fungi [Citation7,Citation22]. Therefore, the precise identification of phytopathogenic species of C. gloeosporioides that cause necrosis in the pericarp tissue and the soft rot mesocarp of fruits is very important [Citation23]. This would allow for the rational and adequate use of fungicides to reduce the incidence and severity of disease [Citation24], and provide the knowledge required for determining the sources of the primary inoculum from which the infection cycle begins in fruits [Citation1].
Here, we demonstrate that anthracnose symptoms are present in unripe avocado fruits in the field in the central part of Mexico, and provide accurate identification of the associated causal agents using morphological and phylogenetic approaches.
2. Material and methods
2.1. Sampling and fungal isolation
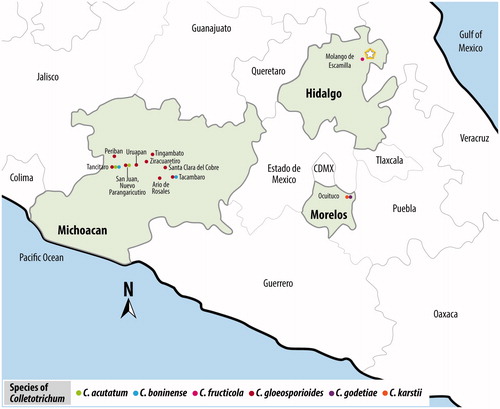
In 2015, isolates of Colletotrichum were obtained from fruits recovered in an orchard located in Molango de Escamilla, state of Hidalgo, Mexico at 1663 m above sea level. Thirty avocado fruits cv. “Hass” exhibiting anthracnose symptoms in the pericarp were sampled for fungal isolation. These symptoms are represented by the formation of cankers as small circular necrotic areas on the fruits. Exocarp tissue fragments, taken from the canker advance zone, were washed in tap water, surface-disinfested in a 1.5% sodium hypochlorite solution for 2 min, and rinsed three times with sterile distilled water (SDW). Small pieces of ∼5 mm2 were dried in a laminar flow chamber on sterile paper towels and placed onto Petri plates containing potato dextrose agar (PDA) medium (Difco, NJ, USA). The plates were incubated at 28 °C in the dark for five days, and then fungal colonies were transferred to PDA plates and maintained at 28 °C in the dark until conidia were observed.
To obtain single conidium isolates, the conidia were harvested by adding 5 mL of SDW to the plates, and the concentration of the suspension was adjusted to 1 × 106 conidia/mL using an hemocytometer (Marienfeld, Germany). A 100-µL aliquot was spread on water agar and incubated at the same temperature as above in the dark for 24 h. A single germinating conidium was transferred to a fresh PDA plate. Twenty-four isolates were obtained and morphologically identified as Colletotrichum based on morphological features of the genus.
2.2. Pathogenicity test
Mycelial discs from a five-day-old actively growing edge colony on PDA medium were transferred to oatmeal agar (OA: 20 g oatmeal, 20 g agar, 1 L distilled water) plates, and scraped onto the surface of the medium to induce sporulation [Citation25]. The plates were incubated at room temperature (24–26 °C) for 10 days.
Twenty-five healthy unripe avocado fruits cv. “Hass” were washed with running tap water and surface-disinfested in 1.5% sodium hypochlorite for 5 min, rinsed three times with SDW, and then dried at room temperature. A conidial suspension (106 conidia/mL) was prepared from each 10-day-old monoconidial Colletotrichum culture by adding 5 mL of SDW to an OA plate. The fruits were inoculated using the non-wound drop inoculation method. Thirty microliters of the conidial suspension was placed on the middle of each fruit. Control fruits were inoculated with 30 µL of SDW. The inoculated samples were incubated in containers at room temperature in normal light for 10 days [Citation26]. Subsequently, the fruits were cut to observe the development of the typical soft rot lesions in the mesocarp. The fungus was re-isolated from the inoculated fruits to fulfill Koch’s postulates, and the identity was determined using a morphological approach.
2.3. DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing
For each Colletotrichum isolate, a small amount of mycelium was scraped from the surface of a 5-day-old culture (grown on PDA at 28 °C) using a sterile 10-µL pipette tip, and genomic DNA was extracted using the 2% CTAB method with slight modifications [Citation27].
The ITS regions of rDNA, including ITS1, 5.8S, and ITS2 (ITS) [Citation28]; actin (ACT), chitin synthase (CHS) [Citation29], glyceraldehyde-3-phosphate dehydrogenase (GAPDH) partial genes [Citation30]; and Apn2-Mat1-2 intergenic spacer and partial mating type Mat1-2 gene (ApMat) [Citation31] of nine selected Colletotrichum sp. obtained in this study were amplified and sequenced. The primers used in this study are shown in .
Table 1. Primer names, primer sequences, and references used in this study.
PCR was performed in 15-µL reaction mixtures containing 0.18 µL of each primer, 0.18 µL of dNTPs, 0.9 U of GoTaq® DNA polymerase (Promega, Madison, WI, USA), and 3 µL of DNA template (20 ng/µL). The PCR protocols were standardized for each gene as follows: ITS, a first step at 95 °C for 4 min, followed by 35 cycles at 95 °C for 1 min, 58 °C for 1 min and 72 °C for 2 min, and a final step at 72 °C for 10 min; ACT, a first step at 95 °C for 8 min, followed by 35 cycles at 95 °C for 15 s, 61 °C for 20 s, 72 °C for 1 min, and a final step at 72 °C for 5 min; CHS, a first step at 95 °C for 4 min, followed by 35 cycles at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, and a final step at 72 °C for 7 min; GAPDH, a first step at 94 °C for 5 min, followed by 35 cycles for 94 °C for 1 min, 56 °C for 75 s, and 72 °C for 75 s, and a final step at 72 °C for 10 min; ApMat, a first step at 95 °C for 5 min, followed by 10 cycles at 95 °C for 30 s, 62 °C for 30 s, and 72 °C for 1 min, followed by 35 cycles at 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 1 min, with a final step at 72 °C for 10 min.
Amplified PCR products were cleaned using ExoSAP-IT® (Affymetrix, Santa Clara, CA, USA) and both strands were directly sequenced using BigDyeTM Terminator v3.1 Cycle Sequencing Kit (Applied BiosystemsTM) in a 3130 Genetic Analyzer Sequencer (Applied BiosystemsTM) at Postgraduate College facilities, Mexico.
2.4. Phylogenetic reconstruction
The DNA sequences obtained from both strands were assembled to obtain consensus sequences for each isolate using BioEdit v7.0.5 [Citation32]. Phylogenetic analysis of the Colletotrichum isolates was performed based on concatenated multi-locus datasets of ITS, ACT, CHS, GAPDH, and ApMAT sequences. Multiple sequences were aligned using the MAFFT v.7 server (http://mafft.cbrc.jp/aligment/server/) under default parameters.
Sequences of Colletotrichum species deposited in the GenBank database belonging to the Musae clade in the C. gloeosporioides species complex [Citation7] were retrieved and included as reference species along with the sequences obtained in this study (). Concatenated alignment was performed using MESQUITE v.3.10 [Citation33], and Bayesian inference (BI) was performed using four Markov Chains Monte Carlo (MCMC) in Mr. Bayes v.3.2.1 software [Citation34]. Maximum-likelihood analysis was performed in raxmlGUI 1.5b2 [Citation35]. Rapid bootstrapping with 1000 iterations was implemented using the general time reversible model with a gamma distribution.
Table 2. Isolates of Colletotrichum spp. used in this study with GenBank accession numbers.
Nucleotide substitution models were determined using MrModeltest v.2.3 [Citation36] for each gene partition and included in the BI. Two analyses of four MCMC chains were run from random trees for 1,500,000 generations and sampled every 1000 generations. The first 25% of generated trees were discarded as the burn-in phase of each analysis and posterior probabilities were determined for the remaining trees. The phylogenetic tree was rooted with the sequence of C. gloeosporioides sensu stricto CBS 123755. Sequences derived in this study were submitted to GenBank (). DNA alignment and phylogenetic trees were deposited in TreeBase under ID study S21326.
2.5. Morphology and cultural characteristics
After phylogenetic reconstruction and identification of the pathogen, morphological analysis was performed. Conidia were harvested from the PDA plates, incubated at 28 °C in the dark, and then mounted onto a microscope slide with clear 1% lactic acid. Mycelia conidial and appressorium formation were produced using a slide culture technique [Citation37,Citation38], and 30 of them were selected for measurement using ImageJ software (https://imagej.nih.gov/ij/) through photographs taken by Infinity 1-2 C implemented on an Olympus BX41 microscope (Tokyo, Japan). The shape and size of 50 conidia were measured with the same procedure. Colony characteristics on PDA were recorded after 7 days.
3. Results
3.1. Monoconidial isolates
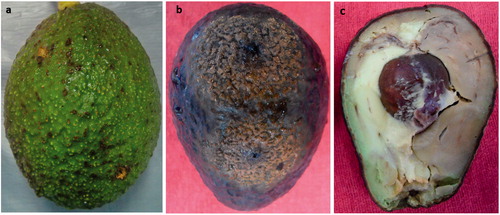
Original field symptoms observed in 30 avocado fruits collected in the central part of Mexico showed necrotic tissue with small dark brown spots on green-unripe fruit exocarps, which coalesced with age (). Twenty-four monoconidial isolates were obtained for further analyses.
3.2. Pathogenicity test
The pathogenicity test was performed with 24 Colletotrichum isolates on unripe fruits of avocado cv. “Hass” to confirm Koch’s postulates. The development of small circular necrotic spots was observed at 10 days after inoculation (DAI), and abundant conidia masses were observed on the surface of the avocado fruits for all isolates inoculated (). When the fruits were cut, a soft rot with dark brown discoloration in the mesocarp was observed at 10 DAI (). Setae were absent. The fungus was re-isolated from the exocarp tissue, as described above, and re-identified morphologically. All of the inoculated isolates were found to be very aggressive because the infection advanced until covering almost 50% of the inoculated fruits.
3.3. Phylogenetic analysis
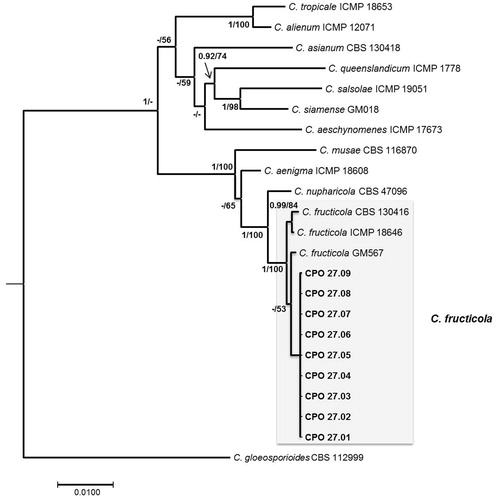
All isolates were similar in the morphological and pathogenicity tests; thus, nine isolates were selected at random for phylogenetic analysis. The combined gene alignment comprised 23 taxa and 1922 characters, including gaps (ITS: 1–507, ACT: 508–729, CHS: 730–1002, GAPDH: 1003–1266, ApMAT: 1267–1922). The best evolution models of each gene were: K80 + I for ITS, HKY for ACT, SYM + I for CHS, HKY + G for GAPDH, and HKY + I for ApMAT. The consensus tree obtained for Bayesian analysis showed that the nine isolates obtained in this study clustered with the C. fructicola clade with a posterior probability of 100%, and differed from the reference isolates. C. fructicola was separated from the most closely related species (C. nupharicola) with 100% support (). All isolates analyzed in this study by a molecular approach were identical for all genes.
Figure 2. Bayesian consensus phylogenetic tree in the Musae clade of the C. gloeosporioides species complex, constructed with concatenated sequences of the ITS region, ACT, CHS, GAPDH, and ApMat with the best evolutionary model applied for each gene. Posterior probabilities ≥0.90 and bootstrap support ≥50% are shown in the nodes (BI/ML). Scale bar: substitutions per site.
3.4. Morphology and cultural characteristics
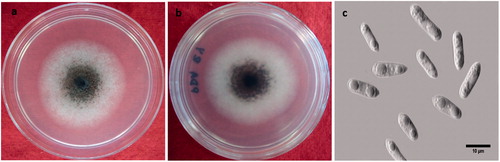
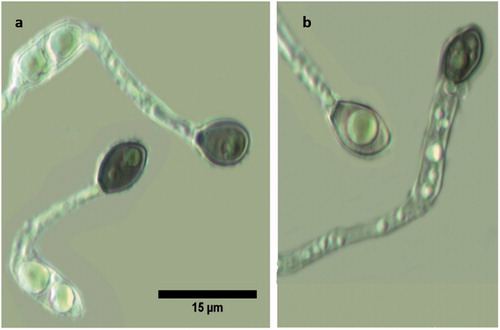
Most of the morphological features coincided with those reported by Prihastuti et al. [Citation39]. For all isolates, the growth of colonies on PDA media showed a dark brown color, becoming gray to dark gray at the center with age (). The aerial mycelium was gray, dense, with visible conidia masses. Acervuli and setae were absent. The reverse side was grayish with a white halo of mycelium (). Conidia were (11.06−) 14 (−16.14) × (3.19−) 4.03 (−4.89) µm (n = 50), one-celled, smooth-walled, hyaline, cylindrical with obtuse to slightly rounded ends; the majority had two rounded guttules (). Conidial and mycelial appressoria formed on slide cultures were (7.68−) 8.46 (−9.62) × (5.65−) 6.13 (−6.74) µm (n = 30), brown to dark brown, ovoid for conidial appressoria and slightly obtuse at the tip, pedicellate, and irregular in shape for mycelial appressoria, often containing one or more guttules ().
4. Discussion
Avocado (P. americana Mill.) is one of the most important fruit crops in Mexico because it is the main export product. In 2013, Mexico ranked first worldwide in production and export with 1,467,837 tons and 563,492 tons, respectively [Citation40]; trades forecast that avocado exports for the marketing year 2016–2017 will increase to over 1.0 million metric tons due to expected strong international demand [Citation41].
One of the main problems faced by producers to obtain fruits with high commercial quality is the incidence of fungal diseases. Anthracnose caused by different species of Colletotrichum represents one of the challenges within production systems. Through amplification of ITS sequences, C. gloeosporioides, C. acutatum, and C. boninense were initially described to be associated with avocado anthracnose fruits in the states of Michoacan and Morelos, Mexico, respectively [Citation15,Citation42]. Furthermore, it has been reported that C. godetiae and C. karstii, which belong to the C. acutatum and C. boninense complexes, affect avocado in the state of Morelos, Mexico [Citation16,Citation17]. In the present study, C. fructicola, a member of the C. gloeosporioides species complex, was found to be a causal agent of anthracnose in avocado fruits collected in the state of Hidalgo, in the central part of Mexico, using morphological and multilocus phylogenetic analyses. Nevertheless, variation in colony morphology (colony color, pigment, sporulation, perfect state, and conidia size) has been reported [Citation43] and sterile isolates of C. fructicola have been encountered [Citation3], suggesting high genetic variation.
Colletotrichum fructicola was proposed as a crop pathogen for the first time by Prihastuti et al. [Citation39] using a polyphasic approach, affecting coffee berries in Thailand. Since then, it has been reported to cause anthracnose, thus representing an important fungal pathogen in several plantations worldwide [Citation6]. Colletotrichum fructicola was the dominant and most aggressive species associated with bitter rot of apple in Uruguay [Citation44] and strawberry in Japan [Citation45]. In China, C. fructicola was reported to cause anthracnose of sandy pear (Pyrus pyrifolia), an economically significant disease in southern China [Citation43], and was the principal fungus of the C. gloeosporioides complex causing citrus anthracnose [Citation46]. Furthermore, C. gloeosporioides, previously reported to cause anthracnose in different crops worldwide, was then reclassified. For example, C. fructicola is a causal agent of strawberry anthracnose in Korea, which was previously reported as C. gloeosporioides [Citation47].
Colletotrichum gloeosporioides sensu stricto as a causal agent of avocado fruits anthracnose has only been reported recently in Turkey [Citation48]. In addition, C. gloeosporioides sensu stricto was not present in tropical fruits tested in Laos and Thailand [Citation49]. Diverse studies reported C. gloeosporioides as a pathogen causing anthracnose in avocado fruits using analyses of the ITS region, in which the isolates were grouped in different clades, suggesting that different species were present [Citation12,Citation15]. Nevertheless, other genes and statistical approaches are needed to determine robust phylogenetic relationships and provide the true species delimitations [Citation23,Citation49]. For example, the intergenic sequence of apn2 and Mat1-2 (ApMat) can improve resolution for the definition of the Colletotrichum species complex compared to other commonly used loci [Citation2,Citation50], and ApMat and GS loci can help to resolve species identification in the C. gloeosporioides complex affecting Camellia [Citation51].
It is possible that C. fructicola is amply distributed in other crops and that diverse species can cause the same disease in avocado because of an ample range host and the high diversity of this genus, which have not been previously investigated in Mexico. However, special attention should be given to this species, because even though anthracnose symptoms in the field were not very evident (), when the fruits were placed in favorable environmental conditions, the fungus was able to colonize almost the entire fruit in a short time () accompanied by mesocarp soft rot (). This observation further indicates the potential aggressiveness of these isolates and the risk that can be introduced in other areas where C. fructicola has not yet been reported to date.
Further research focused on determining the distribution of Colletotrichum species is needed for implementation of specific management strategies based on fungicide applications in each avocado production region, because variation in sensitivity can occur as well as the emergence of fungicide-resistant isolates [Citation52,Citation53]. Furthermore, the emergence of more aggressive species of Colletotrichum highlights the need for regular surveys, particularly when taking into account the fact that most commercial groves are cv. “Hass” monocultures, the effects of climate change, and the adaptation of pathogens to new varieties and ecological niches.
In conclusion, the present results obtained by molecular and morphological techniques as well as pathogenicity tests suggest that C. fructicola is the causal agent of anthracnose and soft rot in avocado fruits cv. “Hass” in the central part of Mexico. This finding is very important because the state of Hidalgo is close to Michoacan, the main avocado-producing state in Mexico, and thus C. fructicola could spread into areas where only C. gloesporioides, C. acutatum, and C. boninense have been reported to date (). Moreover, this species should be included in sensitivity to fungicides tests aimed at maintaining the high quality of avocado fruits, which is essential for the exportation trade.
Acknowledgment
The authors are grateful to Petra Aguirre-Rayo for her technical assistance in the sequencing facilities at the College of Postgraduates.
Disclosure statement
The authors declare no conflict of interest related to any part of the research.
Additional information
Funding
References
- Freeman S, Katan T, Shabi E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Dis. 1998;82:596–605.
- Silva DN, Talhinhas P, Várzea V, et al. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia. 2012;104:396–409.
- Lima NB, Batista MV de A, De Morais MA Jr, et al. Five Colletotrichum species are responsible for mango anthracnose in north-eastern Brazil. Fungal Divers. 2013;61:75–88.
- Cristobal-Martínez AL, Yañez-Morales MJ, Solano-Vidal R, et al. Diversity of Colletotrichum species in coffee (Coffea arabica) plantantions in Mexico. Eur J Plant Pathol. 2017;147:605–614.
- Li H, Zhou G-Y, Liu J-A, et al. Population genetic analyses of the fungal pathogen Colletotrichum fructicola on tea-oil trees in China. PLoS One. 2016;11:e0156841.
- Farr DF, Rossman AY. Fungal database: fungus–host distributions, U.S. National Fungus Collections, ARS, USDA [Internet]. [cited 2017 May 15]. Available from: https://nt.ars-grin.gov/fungaldatabases/
- Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud Mycol. 2012;73:115–180.
- Jayawardena RS, Hyde KD, Damm U, et al. Notes of currently accepted species of Colletotrichum. Mycosphere. 2016;7:1192–1260.
- Diao YZ, Zhang C, Liu F, et al. Colletotrichum species causing anthracnose disease of chili in China. Persoonia. 2017;38:20–37.
- Guarnaccia V, Groenewald JZ, Polizzi G, et al. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia. 2017;39:32–50.
- Dann EK, Ploetz RC, Coates LM, et al. Foliar, fruit and soil borne diseases. In: Schaffer B, Wolstenholme BN, Whiley AW, editors. The avocado, botany, production and uses. London: CAB International; 2013. p. 380–422.
- Sanders GM, Korsten L. A comparative morphological study of South African avocado and mango isolates of Colletotrichum gloeosporioides. Can J Bot. 2003;81:877–885.
- Freeman S, Minz D, Jurkevitch E, et al. Molecular analyses of Colletotrichum species from almond and other fruits. Phytopathology. 2000;90:608–614.
- Damm U, Cannon PF, Woudenberg JHC, et al. The Colletotrichum acutatum species complex. Stud Mycol. 2012;73:37–113.
- Silva-Rojas HV, Ávila-Quezada GD. Phylogenetic and morphological identification of Colletotrichum boninense: a novel causal agent of anthracnose in avocado. Plant Pathol. 2011;60:899–908.
- Hernández-Lauzardo AN, Campos-Martínez A, Velázquez-del-Valle MG, et al. First report of Colletotrichum godetiae causing anthracnose on avocado in Mexico. Plant Dis. 2015;99:555.
- Velázquez-del-Valle MG, Campos-Martínez A, Flores-Moctezuma HE, et al. First report of avocado anthracnose caused by Colletotrichum karstii in Mexico. Plant Dis. 2016;100:534.
- Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246.
- Abang MM, Winter S, Green KR, et al. Molecular identification of Colletotrichum gloeosporioides causing yam anthracnose in Nigeria. Plant Pathol. 2002;51:63–71.
- Chung WH, Ishii H, Nishimura K, et al. Fungicide sensitivity and phylogenetic relationship of anthracnose fungi isolated from various fruit crops in Japan. Plant Dis. 2006;90:506–512.
- Shivas RG, Tan YP. A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Divers. 2009;39:111–122.
- Damm U, Cannon PF, Woudenberg JHC, et al. The Colletotrichum boninense species complex. Stud Mycol. 2012;73:1–36.
- Liu F, Wang M, Damm U, et al. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evol Biol. 2016;16:81.
- Bragança CA, Damm U, Baroncelli R, et al. Species of Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biol. 2016;120:547–561.
- Suzaki K. Improving method to induce sporulation of Colletotrichum gloeosporioides, causal fungus of grape ripe rot. J Gen Plant Pathol. 2011;77:81–84.
- Cai L, Hyde KD, Taylor PWJ, et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009;39:183–204.
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556.
- Templeton MD, Rikkerink EH, Solon SL, et al. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene. 1992;122:225–230.
- Rojas EI, Renher SA, Samuels GJ, et al. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia. 2010;102:1318–1338.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis version 3.10 [Internet]. 2016. Available from: http://mesquiteproject.org
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542.
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12:335–337.
- Nylander JAA. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004.
- Johnston PR, Jones D. Relationship among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia. 1997;89:420–430.
- Yang YL, Liu ZY, Cai L, et al. Colletotrichum anthracnose of Amaryllidaceae. Fungal Divers. 2009;39:123–146.
- Prihastuti H, Cai L, Chen H, et al. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009;39:89–109.
- FAO. FAOSTAT [Internet] [cited 2017 Jan 20]. Available from: http://faostat.fao.org
- Flores D. Greater volume of Mexican avocados to the U. S. market. USDA [Internet]. USA; [cited 2017 Jan 25]. Available from: https://gain.fas.usda.gov/Recent%20GAIN%20Publications/Avocado%20Annual_Mexico%20City_Mexico_11-30-2016.pdf
- Campos-Martínez A, Velázquez-del-Valle MG, Flores-Moctezuma HE, et al. Antagonistic yeasts with potential to control Colletotrichum gloeosporioides (Penz.) & Sacc. and Colletotrichum acutatum J. H. Simmonds on avocado fruits. Crop Prot. 2016;89:101–104.
- Zhang PF, Zhai LF, Zhang XK, et al. Characterization of Colletotrichum fructicola, a new causal agent of leaf black spot disease of sandy pear (Pyrus pyrifolia). Eur J Plant Pathol. 2015;143:651–662.
- Alaniz S, Hernández L, Mondino P. Colletotrichum fructicola is the dominant and one of the most aggressive species causing bitter rot of apple in Uruguay. Trop Plant Pathol. 2015;40:265–274.
- Gan P, Nakata N, Suzuki T, et al. Markers to differentiate species of anthracnose fungi identify Colletotrichum fructicola as the predominant virulent species in strawberry plants in Chiba Prefecture of Japan. J Gen Plant Pathol. 2017;83:14–22.
- Huang F, Chen GQ, Hou X, et al. Colletotrichum species associated with cultivated citrus in China. Fungal Divers. 2013;61:61–64.
- Nam MH, Park MS, Lee HD, et al. Taxonomic re-evaluation of Colletotrichum gloeosporioides isolated from strawberry in Korea. Plant Pathol J. 2013;29:317–322.
- Akgül DS, Awan QN, Güler PG, et al. First report of anthracnose and stem end rot diseases caused by Colletotrichum gloeosporioides and Neofusicoccum australe on avocado fruits in Turkey. Plant Dis. 2016;100:1792.
- Phoulivong S, Cai L, Chen H, et al. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers. 2010;44:33–43.
- Sharma G, Kumar N, Weir BS, et al. The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Divers. 2013;61:117–138.
- Liu F, Weir BS, Damm U, et al. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Pers – Int Mycol J. 2015;35:63–86.
- Afanador-Kafuri L, González A, Gañan L, et al. Characterization of the Colletotrichum species causing anthracnose in Andean blackberry in Colombia. Plant Dis. 2014;98:1503–1513.
- Velho AC, Alaniz S, Casanova L, et al. New insights into the characterization of Colletotrichum species associated with apple diseases in southern Brazil and Uruguay. Fungal Biol. 2015;119:229–244.