Abstract
Rice blast disease, caused by the ascomycete fungus Magnaporthe oryzae, is one of the most important diseases in rice production. PAS (period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, single-minded protein) domains are known to be involved in signal transduction pathways, but their functional roles have not been well studied in fungi. In this study, targeted gene deletion was carried out to investigate the functional roles of the PAS-containing gene MoPAS1 (MGG_02665) in M. oryzae. The deletion mutant ΔMopas1 exhibited easily wettable mycelia, reduced conidiation, and defects in appressorium formation and disease development compared to the wild type and complemented transformant. Exogenous cAMP restored appressorium formation in ΔMopas1, but the shape of the restored appressorium was irregular, indicating that MoPAS1 is involved in sensing the hydrophobic surface. To examine the expression and localization of MoPAS1 in M. oryzae during appressorium development and plant infection, we constructed a MoPAS1:GFP fusion construct. MoPAS1:GFP was observed in conidia and germ tubes at 0 and 2 h post-infection (hpi) on hydrophobic cover slips. By 8 hpi, most of the GFP signal was observed in the appressoria. During invasive growth in host cells, MoPAS1:GFP was found to be fully expressed in not only the appressoria but also invasive hyphae, suggesting that MoPAS may contribute to disease development in host cells. These results expand our knowledge of the roles of PAS-containing regulatory genes in the plant-pathogenic fungus M. oryzae.
Keywords:
1. Introduction
Fungi have evolved to sense ambient environmental signals, such as air, light, and host surface features, for appropriate development and survival [Citation1]. For example, G-protein-coupled receptors (GPCRs), one of the largest families of transmembrane receptors, are involved in sensing environmental signals and activating signal transduction pathways [Citation2]. The PAS (period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, single-minded protein) domain is involved in sensing signals, and is present in many signaling proteins, such as histidines (HKs) and serine/threonine (S/T) kinases, as well as voltage-gated ion channels [Citation3,Citation4]. The PAS domain was first discovered in Drosophila proteins, and has since been found in organisms ranging from bacteria to humans [Citation3,Citation5]. In bacteria, PAS domains are known to associate with histidine kinase (HK) sensor proteins [Citation6]. HKs were initially believed to exist only in bacteria, but in the 1990s it was revealed that HKs also regulate essential processes in fungi [Citation7]. In addition to their sensory role, PAS domains also modulate protein–protein interactions in response to stimuli, which allows for complex cellular signaling networks [Citation8].
PAS domains are found in previously characterized regulators in some fungi. For example, PAS domains are present in the serine/threonine protein kinases PSK1 and PSK2 in yeast [Citation9]. In yeast, PSK1 regulates superoxide dismutase-1 (SOD1) to protect against oxidative stress, and PSK2 acts as a nutrient-sensing kinase [Citation10]. In the filamentous fungus Neurospora crassa, DCC-1, a putative HK, is involved in the regulation of conidiation, perithecial development, and carotenogenesis [Citation11]. In the ascomycete fungus Aspergillus nidulans, phytochrome FphA regulates sexual development and mycotoxin formation [Citation12]. These studies suggest important roles for PAS-containing proteins in diverse aspects of fungal development.
Magnaporthe oryzae is a filamentous plant pathogen that causes rice blast disease. This fungus has been used as a model organism for studying the molecular basis for fungal development and pathogenicity [Citation13]. On rice leaves, conidia germinate and develop specialized infection structures called appressoria in response to host signaling factors. Sensing host signaling factors and activating signaling pathways are known to be necessary for M. oryzae to infect rice. cAMP-dependent protein kinase signaling is needed for appressorium formation in response to plant signaling factors in M. oryzae [Citation14,Citation15]. Pth11, a GPCR, is also known to be important in hydrophobic surface sensing for appressorium formation [Citation16,Citation17]. Recently, HKs were functionally characterized in M. oryzae [Citation18]. In this previous study, the deletion mutant ΔMohik5 was unable to form appressoria and was therefore completely nonpathogenic.
Previously, we identified conidiation-related genes from microarray analysis, among which attention has been paid to MoPAS1 (MGG_02665) due to potential important roles in fungal development and pathogenicity [Citation19]. We generated a targeted gene deletion mutant for MoPAS1, named as ΔMopas1. Deletion of MoPAS1 resulted in pleiotropic defects in conidiation, surface hydrophobicity of mycelia, appressorium development, and disease development. The defect of ΔMopas1 in appressoria formation on hydrophobic coverslips was restored by cAMP treatment, which suggested that MoPAS1 plays an important role in surface sensing. Collectively, the results showed that MoPAS1 plays critical roles in preinfection-associated developmental stages and disease development in M. oryzae.
2. Materials and methods
2.1. Phylogenetic analysis
All sequence information was obtained from the Magnaporthe genome database of the Broad Institute (http://www.broadinstitute.org), the Comparative Fungal Genomics Platform (CFGP) online database (http://cfgp.snu.ac.kr), and the BLAST program provided by the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov). Sequence alignment was performed using ClustalW program in MEGA 6.0 [Citation20,Citation21]. Phylogenetic analysis was performed using the neighbor-joining method (1000 bootstrap replicates). Domain structure analysis was performed using InterPro Scan v53.0 (http://www.ebi.ac.uk/interpro/) [Citation22]. Domain structure analysis was performed using InterPro Scan (http://www.ebi.ac.uk/interpro/) [Citation22]. The primers used in this study () were designed using the PrimerQuest Design Tool (http://sg.idtdna.com/site), and purchased from Bioneer (Daejeon, Korea).
Table 1. Primers used in this study.
2.2. Fungal strains, culture conditions, and phenotypic characterization
The M. oryzae wild-type KJ201 and transformants used in this study were cultured on oatmeal agar (OMA, 50 g of oatmeal and 25 g of agar per liter) or V8 juice agar (V8A, 80 mL of V8 juice, 310 μL of 10 N NaOH solution, and 15 g of agar per liter) at 25 °C under continuous light. Conidiation of M. oryzae was measured by counting the number of conidia harvested from 7-d-old OMA plates using a hemocytometer. For microscopic observation of conidiation, mycelial agar plugs from 4-d-old OMA plates were transferred on slide glasses using a cork borer and covered with cover slips. After 1 d, conidiation was observed under a light microscope. To measure conidial germination and appressorium formation, we collected conidia from 7-d-old OMA plates and placed drops (20 µL) of conidial suspensions (5 × 104 conidia/mL) on hydrophobic cover slips and incubated them in a moistened box. Germination was examined by counting the number of germinated conidia from 100 conidia per coverslip after 2 h and appressorium formation was measured after 16 h. For cAMP assays, we prepared cAMP solution (10 mM) in sterile distilled water (SDW), and cAMP solutions were added to the conidial suspensions to a final concentration of 5 mM after a 2-h incubation. For surface hydrophobicity assays, drops (10 μL) of SDW or 0.2% SDS were placed on wild-type and mutant strains grown on OMA plates for 10 d. Experiments were repeated three times with three replicates for each repeat. All data were processed using the SigmaStar statistical software package (SPSS Science, Chicago, IL).
2.3. Fungal transformation for gene knock-out and complementation
Approximately 1.0-kb flanking regions of MoPAS1 were amplified using primers MoPAS1_5F/5R and MoPAS1_3F/3R from wild-type genomic DNA. The hygromycin phosphotransferase gene (HPH) cassette was amplified from the plasmid pBCATPH using HPH_F/R primers. The amplified fragments were fused by double-joint PCR [Citation23]. The fused construct was amplified using MoPAS_NF/NR primers. The amplified constructs were transformed to wild-type protoplasts using the polyethylene glycol (PEG)-mediated transformation method [Citation24]. Hygromycin-resistant transformants were screened on transformation agar (TB3, 200 g of sucrose, 3 g of yeast extract, 3 g of casamino acids, 10 g of glucose, and 8 g of agar per liter) containing Hygromycin B (200 μg/mL). Targeted gene deletion was confirmed using Southern blot hybridization and RT-PCR [Citation25,Citation26]. For complementation, MoPAS1 with native promoters was amplified from wild-type genomic DNA using MoPAS1_cmF/R primers and co-transformed to ΔMopas1 protoplasts with plasmid pII99, which contains a geneticin-resistance cassette [Citation27].
2.4. RNA isolation, RT-PCR, and gene expression analysis
Total RNA was extracted from frozen fungal tissues using the Easy-Spin Total RNA Extraction Kit (Intron Biotechnology, Seongnam, Korea) according to the manufacturer’s instructions. For reverse transcription PCR (RT-PCR), 5 µg of total RNA was reverse transcribed into first-strand cDNA using oligo (dT) primers with the SuperScript III First-Strand Synthesis System Kit (Invitrogen Life Technologies, Carlsbad, CA). RT-PCR was performed in 20-μL reaction mixtures using Pfu polymerase (Elpis, Daejeon, Korea). Cycling conditions were as follows: 3 min at 95 °C (1 cycle) followed by 20 s at 95 °C, 20 s at 57 °C, and 30 s at 72 °C (30 cycles). The β-tubulin (MGG_00604) gene was used as a control. Quantitative real-time PCR was performed on the StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA) using HIPI Real-Time PCR 2× Master Mix (SYBR Green) (Elpis), and the relative abundance of MPG1 and MHP1 transcripts was measured as described previously [Citation25]. The experiment was performed with three independent pools of tissues in two sets of experimental replicates.
2.5. Pathogenicity assays
For spray assays, 3-week-old rice seedlings (cv. Nakdongbyeo) were sprayed with conidial suspensions (5 × 105 per mL) and incubated in a dew chamber 25 °C for 7 d [Citation28]. For mycelial infection assays, mycelia agar plugs were placed on 4-week-old rice leaves cut into 8-cm lengths and incubated in a moistened box at room temperature for 7 d. To determine the penetration and infection ability of conidia, conidial suspensions (1 × 105 conidia/mL) were inserted into 4-week-old rice sheath tissue or placed on onion epidermis and incubated in a moistened box. Microscopic observation was performed at 24 and 48 h post-infection (hpi). Experiments were repeated three times with three replicates for each repeat.
2.6. Localization of MoPAS1:GFP fusion protein
Overlap cloning was performed to generate the MoPAS1:GFP vector (Han et al., 2015). A 4.4-kb fragment including a 2.4-kb ORF and 2.0-kb native promoter of MoPAS1 was amplified using GFP_F/R primers (). A 5.1-kb fragment from the plasmid pIGPAPA was amplified using pIGPAPA_F/R primers. The amplified 2.0-kb and 5.1-kb fragments were cloned using the Overlap DNA Cloning Kit (Elpis Biotech, Korea), and the fused vector was transformed into wild-type [Citation25]. Fluorescence microscopy images of MoPAS1:GFP localization were acquired using a Carl Zeiss Axio Image A2 microscope (Carl Zeiss Microscope Division, Oberkochen, Germany).
3. Results
3.1. Phylogenetic analysis of MoPAS1
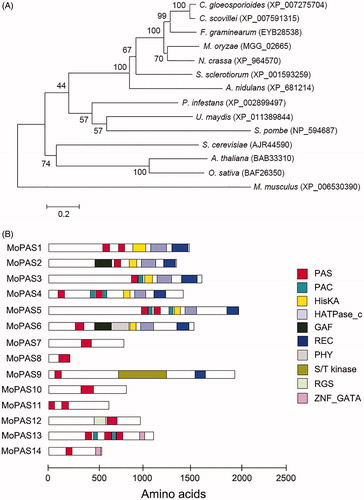
The MoPAS1 (MGG_02665) was predicted to encode a 1481 amino acids. To analyze genetic relationship of MoPAS1 with other organisms, a phylogenetic tree was constructed based on alignments of MoPAS1 amino acid sequences and related proteins in other organisms (). In the phylogenetic tree, MoPAS1 was closely related to proteins in the subphylum Pezizomycotina filamentous fungi Colletotrichum gloeosporioides, C. scovillei, Fusarium graminearum, M. oryzae, Neurospora crassa, Sclerotinia sclerotiorum, and Aspergillus nidulans but distantly related to proteins in the subphylum Saccharomycotina or phylum Basidiomycota, which suggests that PAS1 proteins have evolved in a lineage-specific manner. Next, we searched for PAS domains in the fungal database (fungiDB.org) and found several proteins containing PAS domains. Fourteen proteins containing PAS domains were found in N. crassa and Fusarium graminearum, whereas fewer were found in Cryptococcus neoformans (nine), Aspergillus fumigatus (seven), Schizosaccharomyces pombe (six), and Saccharomyces cerevisiae (three) (data not shown). A total of 14 proteins were found to have PAS domains in M. oryzae strain KJ201 (). These proteins were named MoPAS1–MoPAS22. Each protein had one to three copies of the PAS domain, and proteins ranged from 225 to 1994 amino acids in length. In MoPAS1, there were two PAS domains, a HisKA (HK phosphoacceptor domain; IPR003661) domain, a HATPase_C (HK-like ATPase; IPR003594) domain, and a REC (signal receiver domain; IPR001789) domain. Among the 14 proteins containing PAS domains from M. oryzae strain KJ201, PAC (C-terminal to PAS motifs; IPR001610), HisKA, HATPase_C, GAF (domain present in phytochrome and cGMP-specific phosphodiesterases; IPR003018), REC, PHY (phytochrome; IPR001294), S/T kinase (serine/threonine kinase; IPR008271), RGS (regulator of G protein signaling; IPR016137) and ZNF_GATA (zinc finger, GATA-type; IPR000679) domains were present (). In summary, PAS domains are present in the regulatory proteins of M. oryzae strain KJ201.
Figure 1. Phylogenetic analysis of MoPAS1. (A) Phylogenetic tree analysis of MoPAS1. A neighbor-joining tree of the amino acid sequence of PAS proteins. Numbers at nodes represent bootstrap values calculated from 1000 replicates. The scale bar indicates the number of amino acid differences per site. Sequence information was taken from the NCBI protein database. GenBank accession number for each protein is followed by species; (B) Predicted domain structure of MoPAS proteins. Sequence information was taken from the Magnaporthe genome database of the Broad Institute, and domain structure analysis was performed using InterPro Scan (http://www.ebi.ac.uk/interpro/). PAS (period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, single-minded protein; IPR0000014), PAC (C-terminal to PAS motifs; IPR001610), HisKA (signal transduction histidine kinase; IPR003661), HATPase_C (histidine kinase-like ATPase; IPR003594), GAF (domain present phytochrome and cGMP-specific phosphodiesterases; IPR003018), REC (signal receiver domain; IPR001789), PHY (phytochrome; IPR001294), S/T kinase (serine/threonine kinase; IPR008271), RGS (regulator of G protein signaling; IPR016137), ZNF_GATA (zinc finger, GATA-type; IPR000679).
3.2. Targeted deletion of MoPAS1
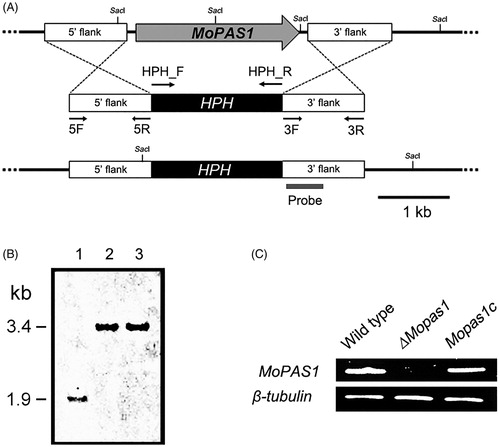
To investigate the functional roles of MoPAS1, we generated a gene deletion mutant, ΔMopas1 (). The deletion construct was amplified by double-joint PCR. The construct is illustrated in . The construct was used to transform protoplasts of M. oryzae strain KJ201 by a PEG-mediated transformation method. More than 100 putative deletion transformants were grown on TB3 agar containing 200 μg/mL hygromycin B, and two candidates were selected by PCR screening using MoPAS1_SF/SR primers (). Finally, ΔMopas1 was selected based on Southern blot assay results. As illustrated in , genomic DNA of the transformants was digested using SacI and hybridized with the probe. Southern blot analysis revealed that a 1.9-kb band was detected in the wild-type and a 3.4-kb band was detected in knock-out mutants (). Next, we confirmed the expression of MoPAS1 by RT-PCR (). The expression of MoPAS1 was completely abolished in ΔMopas1, and was restored in Mopas1c ().
Figure 2. Gene deletion construct and identification of deletion mutants. (A) MoPAS1 was deleted through the targeted gene replacement method; (B) Confirmation of MoPAS1 deletion using Southern blot analysis. DNA samples were digested with SacI. A 1.9-kb band from wild-type KJ201 and a 3.4-kb band from knock-out mutants were produced. Lane 1, wild-type; lane 2–3, knock-out mutants; (C) Confirmation of MoPAS1 deletion and complemented transformant Mopas1c using RT-PCR.
3.3. MoPAS1 is essential for conidiation and appressorium development
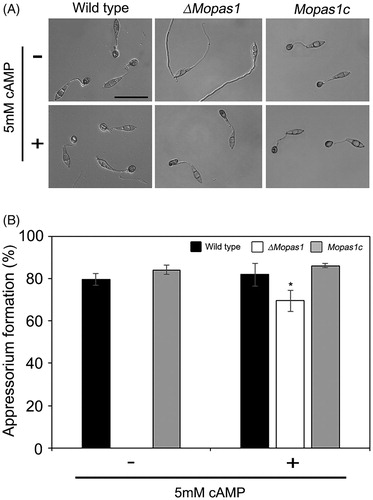
Conidia are important for disease dissemination in M. oryzae. To investigate the functional role of MoPAS1 in conidiation, we assessed the conidiation of ΔMopas1 on OMA media. ΔMopas1 exhibited significantly reduced conidiation on OMA media (). At 7 d after inoculation on OMA, the wild-type strain produced approximately 45 × 104 conidia/mL, whereas ΔMopas1 only produced 26 × 104 conidia/mL. This result means that MoPAS1 is involved in conidiation. To investigate whether conidia of ΔMopas1 are normal in appressorium formation, we incubated 20 µL of conidial drops (5 × 104 conidia/mL) on hydrophobic coverslips. The conidia of wild-type germinated and formed melanized appressoria. ΔMopas1 conidia also germinated on hydrophobic coverslips like wild-type but was unable to develop appressoria, which indicates that MoPAS1 is involved in appressorium formation (). We then investigated whether the defect of ΔMopas1 in appressorium formation is related to cAMP-dependent signal. We added exogenous cAMP solutions on the conidial drops on hydrophobic coverslips. The addition of exogenous cAMP restored appressorium formation in ΔMopas1. However, the restored appressoria of ΔMopas1 were abnormally shaped, indicating that MoPAS1 is involved in surface recognition and the morphogenesis of infection structures.
Figure 3. Conidiation and conidiophore differentiation. (A) Microscopic visualization of conidia and conidiophores in the wild-type strain, ΔMopas1, and Mopas1c; (B) Quantitative measurement of conidia. Each strain was grown on OMA for 7 d, and conidia were counted using a hemocytometer. Asterisks represent significant differences from the wild-type strain (Tukey’s test, p < .05).
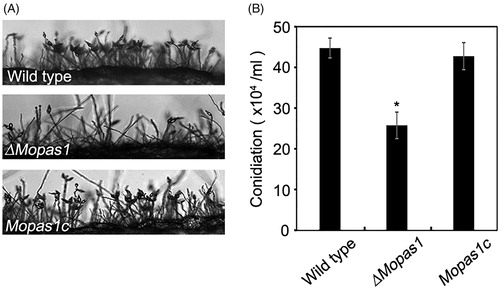
Figure 4. Appressorium formation. (A and B) Conidial suspensions of the wild-type strain, ΔMopas1, and Mopas1c were incubated for 12 hpi on hydrophobic surfaces. For cAMP assays, conidial suspensions of each strain were incubated for 12 hpi on hydrophobic surfaces. cAMP solutions were added to the conidial suspensions to a final concentration of 5 mM at 2 hpi. Asterisks represent significant differences from the wild-type strain (Tukey’s test, p < .05). Bar = 50 μm.
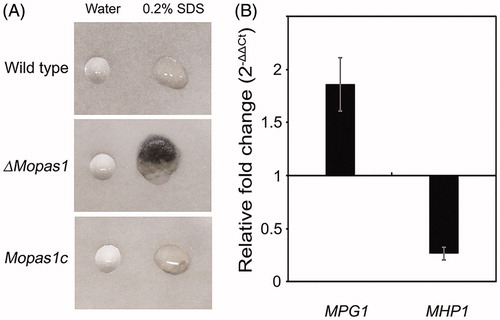
3.4. MoPAS1 is indispensable for surface hydrophobicity
Hydrophobins are low-molecular-weight proteins known to confer hydrophobicity in fungal conidia and hyphae [Citation29]. We next examined surface hydrophobicity by dropping water or SDS on the colony surface of the wild-type strain and ΔMopas1 (). When water drops were placed on wild-type and ΔMopas1, the drops were suspended on the aerial surface of both strains (). However, when the detergent agent SDS was dropped onto the strains, SDS drops immediately soaked into the aerial structures of ΔMopas1 but remained on the surface of the wild-type strain. To investigate whether the defect of hydrophobicity in ΔMopas1 was related to hydrophobins, we measured the mRNA levels of hydrophobin genes MPG1 and MHP1 in ΔMopas1 using qRT-PCR (). MPG1 expression increased in ΔMopas1 compared with the wild-type, whereas MHP1 expression decreased (). This result suggests that MoPAS1-mediated regulation is involved in MPG1 induction and MHP1 repression to enable hydrophobicity in fungal tissue.
Figure 5. Surface hydrophobicity assessment of mycelia. (A) Drops (10 μL) of water or 0.2% SDS were placed on the colony surface of each strain grown on oatmeal agar plates for 10 d; (B) Quantitative measurement of MPG1 and MHP1 gene expression in the mycelia of ΔMopas1 by qRT-PCR, normalized to β-tubulin and expressed relative to expression in the mycelia of the wild-type strain.
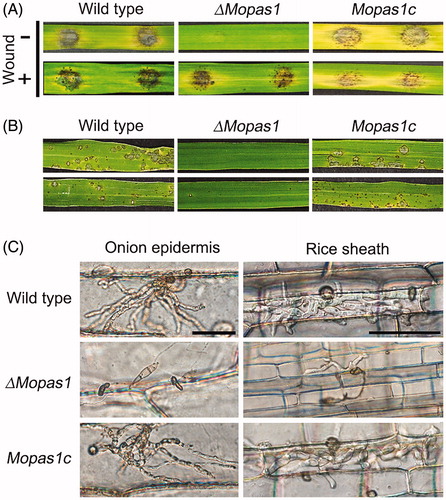
3.5. MoPAS1 is important for fungal pathogenicity
To determine fungal pathogenicity, rice leaves were inoculated using mycelial agar plugs grown on OMA (). Wild-type developed disease lesions on rice leaves, but ΔMopas1 failed to generate disease lesions (). We repeated this experiment with leaves wounded by puncture using a needle. On wounded leaves, mutants formed disease lesions similar to the wild-type (). When conidial suspensions of strains were sprayed on rice leaves, wild-type created disease lesions, whereas ΔMopas1 did not (). To investigate whether this loss of pathogenicity was due to appressorium-mediated penetration, we performed penetration assays of rice sheath and onion epidermis (). In both rice sheath and onion epidermis, abnormal appressoria were observed when ΔMopas1 was inoculated, and most of these abnormal appressoria formed could not penetrate the rice sheath or onion epidermis. The invasive hyphal growth was also significantly attenuated compared to the wild-type (). In contrast, the wild-type and Mopas1c developed normal appressoria and were able to penetrate the plants. These results suggest that MoPAS1 plays an important role in appressorium-mediated penetration in M. oryzae.
Figure 6. Pathogenicity assays. (A) Leaves of the rice cultivar Nakdongbyeo were inoculated with mycelial agar plugs and incubated for 7 d; (B) Conidial suspensions were sprayed onto rice leaves and incubated for 7 d; (C) Conidial suspensions of each strain were inserted into rice sheath and onion epidermis and incubated for 48 h. Bar = 50 μm.
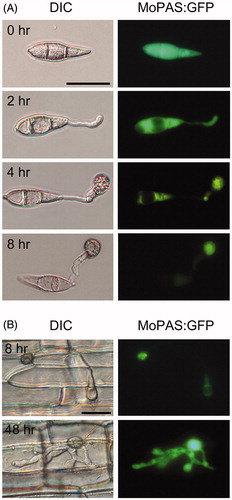
3.6. Subcellular localization of MoPAS1 protein
To investigate the localization of MoPAS1, we constructed MoPAS1:GFP and inserted it into the protoplasts of wild-type. On hydrophobic cover glasses, which induces appressorium formation of M. oryzae, the MoPAS1:GFP fusion proteins were initially distributed uniformly in the cytoplasm of conidia (). As conidia germinates, the MoPAS1:GFP signal was observed in the germ tube. After 8 h, MoPAS1:GFP fusion proteins in conidia and germ tubes disappeared and were only observed inside appressoria. On rice sheath cells, MoPAS1:GFP fusion proteins were intensely localized inside appressoria after 8 h, and uniformly distributed in intracellular invasive hyphae ().
Figure 7. Localization of MoPAS1:GFP in Magnaporthe oryzae strain KJ201 on hydrophobic coverslips or in plants. (A) During appressorium development on hydrophobic coverslips, MoPAS1:GFP was localized in the conidia, germ tubes, and appressoria; (B) In planta observations in rice sheath. MoPAS1:GFP were uniformly localized in invasive hypha. Bar = 20 μm.
4. Discussion
We searched for PAS domains in a fungal database (fungiDB.org) and identified a total of 14 genes containing PAS domains in the M. oryzae strain KJ201 genome. Among these genes, six were identified to contain HisKA (signal transduction HK) domains (). HK is a sensor protein that is typically found in fungi as a hybrid protein including HisKA and REC (signal receiver) domains [Citation30]. It has been reported that PAS-containing HKs are important for infection in plant-pathogenic fungi [Citation18]. Other genes also contained signal transduction domains, including RGS (regulator of G protein signaling) and S/T (serine/threonine kinase) domains. The RGS domain is a conserved domain found in G proteins that regulates G protein-mediated cell signaling [Citation31,Citation32]. For example, in fungi, GPCRs trigger signaling cascades such as the cyclic AMP-dependent protein kinase (cAMP-PKA) or mitogen-activated protein kinase (MAPK) cascades, leading to the activation of cellular processes such as vegetative growth, morphogenesis, and appressorium formation [Citation33]. These results suggest the importance of PAS domains as diverse regulators of cellular functions in fungi.
In the current study, we investigated the functional roles of MoPAS1 (MGG_02665) using a targeted gene-deletion strategy. ΔMopas1 did not form appressoria on hydrophobic surfaces, but this defect was restored when 5 mM cAMP was added to ΔMopas1 (). A previous study revealed the importance of cAMP in the infection-related development of M. oryzae [Citation34], and found that the addition of cAMP induced appressoria formation by M. oryzae on a non-inductive surface. Another study revealed that a protein kinase, MoYAK1, is associated with the cAMP pathway [Citation25]. Consistent with the results of the present study, ΔMoyak1 did not form appressoria on hydrophobic surfaces, but the addition of cAMP induced appressoria formation. Together, the results of the current study and previous reports suggest that the defect in preinfection-associated developmental stages in M. oryzae may be related to the signaling pathway of cAMP-mediated appressorium formation.
ΔMopas1 produced significantly fewer lesions on rice leaves, and the conidia of ΔMopas1 formed irregularly shaped appressoria on rice sheath and onion epidermis (). A recent study highlighted the importance of HKs in fungal pathogenicity in M. oryzae [Citation18]. In this previous study, 10 putative HK-encoding genes were functionally characterized in M. oryzae strain 70–15. In these 10 putative genes, the PAS-containing proteins Hik2p, Hik4p, Hik5p, and Hik8p were involved in pathogenicity on rice cultivar CO-39. However, another PAS-containing protein, Hik9p, was not involved in pathogenicity. ΔMohik9 (MGG_02665) caused disease lesions in the rice cultivar CO-39 similar to those of the wild-type 70-15 strain. These results differed from those of the present study, which may be attributable to the different wild-type strains of M. oryzae used, or to differences among rice cultivars. We found that MoPAS1:GFP fusion protein was localized in the cytosol of conidia, germ tubes, and appressoria during appressorium development on a hydrophobic coverslip. The fusion protein was expressed during plant infection and intensely concentrated in the penetrating appressorium (). These results support the conclusion that MoPAS1 plays an important role in disease development in M. oryze KJ201.
Hydrophobins confer surface hydrophobicity to fungal conidia and hyphae by coating external surfaces [Citation29]. The mycelial surface of ΔMopas1 was detergent-wettable, unlike that of the wild-type, and the relative expression levels of the hydrophobin-containing genes MPG1 and MHP1 were also altered compared to those of the wild-type (). It has been reported that the deletion of MHP1 affects conidiation, appressorium development, and pathogenicity [Citation35]. It has also been reported that MoYAK1, an M. oryzae protein kinase gene, is involved in the regulation of surface hydrophobicity [Citation25]. In ΔMoyak1, the expression of MPG1 and MHP1 was significantly altered. ΔMoyak1 exhibited an easily wettable phenotype and reduced conidiation compared to the wild-type, as well as defective appressorium formation. The defect in appressorium formation was restored by the addition of 5 mM cAMP. These results suggest that MoPAS1 is involved in the regulation of MPG1 and MHP1 and plays an important role in surface hydrophobicity for M. oryzae. We suggest that MoPAS1 is involved in various cellular functions associated with conidiation, surface hydrophobicity, and disease development in M. oryzae strain KJ201. Further studies on the other members of the PAS domain family will help to characterize fungal development and pathogenicity in response to diverse environmental signals.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- van der Does HC, Rep M. Adaptation to the host environment by plant-pathogenic fungi. Ann Rev Phytopathol. 2017;55(1):427–450.
- Kulkarni RD, Thon MR, Pan H, et al. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 2005;6(3):R24.
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63(2):479–506.
- Vreede J, van der Horst MA, Hellingwerf KJ, et al. PAS domains. Common structure and common flexibility. J Biol Chem. 2003;278(20):18434–18439.
- Rojas-Pirela M, Rigden DJ, Michels PA, et al. Structure and function of per-ARNT-sim domains and their possible role in the life-cycle biology of Trypanosoma cruzi. Mol Biochem Parasitol. 2018;219:52–66.
- Gilles-Gonzalez M, Gonzalez G. Signal transduction by heme-containing PAS-domain proteins. J Appl Physiol. 2004;96(2):774–783.
- Herivaux A, So YS, Gastebois A, et al. Major sensing proteins in pathogenic fungi: the hybrid histidine kinase family. PLoS Pathog. 2016;12:e1005683.
- Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17(10):1282–1294.
- Grose JH, Smith TL, Sabic H, et al. Yeast PAS kinase coordinates glucose partitioning in response to metabolic and cell integrity signaling. EMBO J. 2007;26(23):4824–4830.
- Huang M, Xu Q, Mitsui K, et al. PSK1 regulates expression of SOD1 involved in oxidative stress tolerance in yeast. FEMS Microbiol Lett. 2014;350(2):154–160.
- Barba-Ostria C, Lledías F, Georgellis D. The Neurospora crassa DCC-1 protein, a putative histidine kinase, is required for normal sexual and asexual development and carotenogenesis. Eukaryot Cell. 2011;10(12):1733–1739.
- Purschwitz J, Muller S, Kastner C, et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol. 2008;18(4):255–259.
- Dean RA, Talbot NJ, Ebbole DJ, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434(7036):980–986.
- Kang SH, Khang CH, Lee YG. Regulation of cAMP-dependent protein kinase during appressorium formation in Magnaporthe grisea. FEMS Microbiol Lett. 1999;170(2):419–423.
- Lee YH, Dean RA. Hydrophobicity of contact surface induces appressorium formation in Magnaporthe grisea. FEMS Microbiol Lett. 1994;115(1):71–75.
- Liu W, Zhou X, Li G, et al. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 2011;7(1):e1001261.
- Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7(3):185–195.
- Jacob S, Foster AJ, Yemelin A, et al. Histidine kinases mediate differentiation, stress response, and pathogenicity in Magnaporthe oryzae. Microbiologyopen. 2014;3(5):668–687.
- Kim KS, Lee YH. Gene expression profiling during condiation in the rice blast pathogen Magnaporthe oryzae. PLoS One. 2012;7(8):e43202.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680.
- Mulder NJ, Apweiler R, Attwood TK, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2004;33(Database issue):D201–205.
- Yu JH, Hamari Z, Han KH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41(11):973–981.
- Sweigard JA, Chumley FG, Valent B. Cloning and analysis of CUT1, a cutinase gene from Magnaporthe grisea. Mol Gen Genet. 1992;232(2):174–182.
- Han JH, Lee HM, Shin JH, et al. Role of the MoYAK1 protein kinase gene in Magnaporthe oryzae development and pathogenicity. Environ Microbiol. 2015;17(11):4672–4689.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989.
- Fu T, Kim JO, Han JH, et al. A small GTPase RHO2 plays an important role in pre-infection development in the rice blast pathogen Magnaporthe oryzae. Plant Pathol J. 2018;34(6):470.
- Park J, Kong S, Kim S, et al. Roles of forkhead-box transcription factors in controlling development, pathogenicity, and stress response in Magnaporthe oryzae. Plant Pathol J. 2014;30(2):136–150.
- Bayry J, Aimanianda V, Guijarro JI, et al. Hydrophobins—unique fungal proteins. PLoS Pathog. 2012;8(5):e1002700.
- Li D, Agrellos OA, Calderone R. Histidine kinases keep fungi safe and vigorous. Curr Opin Microbiol. 2010;13(4):424–430.
- Li X, Zhong K, Yin Z, et al. The seven transmembrane domain protein MoRgs7 functions in surface perception and undergoes coronin MoCrn1-dependent endocytosis in complex with Gα subunit MoMagA to promote cAMP signaling and appressorium formation in Magnaporthe oryzae. PLoS Pathog. 2019;15(2):e1007382.
- Xu X, Li G, Li L, et al. Genome-wide comparative analysis of putative Pth11-related G protein-coupled receptors in fungi belonging to Pezizomycotina. BMC Microbiol. 2017;17(1):166.
- Sabnam N, Roy Barman S. WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet Biol. 2017;105:37–51.
- Lee YH, Dean RA. cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell. 1993;5(6):693–700.
- Kim S, Ahn IP, Rho HS, et al. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol. 2005;57(5):1224–1237.
