Abstract
The Mount Juwang was designated as a national park in 1976 because of its unique bedrock geology. Although Juwang National Park has maintained its natural ecosystem well, few macrofungal surveys have been carried out. As a part of a project supported by the National Institute of Biological Resources (NIBR) for the discovery of indigenous fungal species, we surveyed the mushrooms in Juwang National Park from 2018 to 2019. The macrofungi were identified based on morphological and molecular analyses. Among these fungi, five specimens were identified as species previously unrecorded in Korea: Calocybe decolorata, Crepidotus brunnescens, Mycena pearsoniana, Psathyrella phegophila, and P. sulcatotuberculosa. Three of these species are known to be rare in the world: Crepidotus brunnescens, P. phegophila, and P. sulcatotuberculosa. In this study, we provide detailed morphological descriptions of the five unrecorded species from Mt. Juwang in Korea.
1. Introduction
Mount Juwang is located in Gyeongsangbuk-do, Korea. The mountain has a specific bedrock geology that is rare in Korea [Citation1]. Mt. Juwang was designated as a national park in 1976. Thus, the biodiversity of Mt. Juwang has been maintained despite the rapid development of the surrounding area. A total of 503 macrofungi were reported in Juwangsan National Park by 2017 [Citation2].
DNA sequence-based identification is a useful and rapid method for identifying novel or unrecorded fungi [Citation3]. The DNA analysis is performed with the internal transcribed spacer (ITS) region as the barcode maker for fungi [Citation4]. Although only using the ITS region shows low species resolution for some fungi, it is still suitable for the identification of most macrofungi. Recently, publications about the identification of unrecorded fungi in China and Korea with a DNA sequence-based approach have rapidly increased [Citation5–7].
As macrofungi have significant ecological and economic impacts, the National Institute of Biological Resources (NIBR) has steadily performed various projects to uncover indigenous fungal species. As a part of the project, we surveyed the macrofungal diversity in Juwang National Park from 2018 to 2019 to discover macrofungi that had not been recorded in Korea. During the investigation, we found five species that are novel in Korea. In this study, we provided detailed morphological descriptions and phylogenetic identities.
2. Materials and methods
2.1. Sampling
The investigation of macrofungal diversity took place in Juwang National Park in 2018 and 2019. Three different sites within the mountains were selected, and the locations are presented in . We sampled the mushrooms and desiccated the samples in a drying oven at 60 °C for 24 h. Subsequently, the samples were placed in a freezer at −20 °C for 3 days to eliminate the vermin. The treated specimens were stored in zipper bags with silica gel.
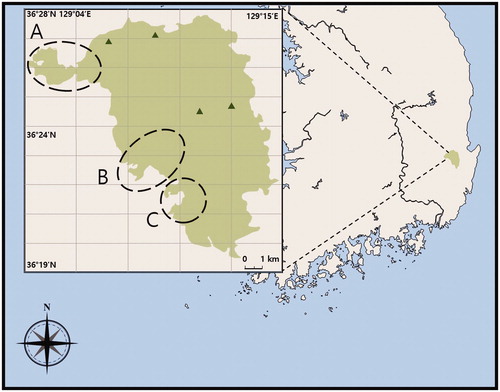
Figure 1. The location of Mount Juwang is presented by green color. The long dashed circles are indicated around the Dalgi Falls (A), Yongchu Falls (B), and Jusan Reservoir (C) in Mountain Juwang. The Calocybe decolorata, Crepidotus brunnescens, and Mycena pearsoniana were collected around the Dalgi Falls (A), and Psathyrella phegophila was around from Jusan Reservoir (C), and P. sulcatotuberculosa was around from Yongchu Falls (B).
2.2. Molecular approach
DNA extraction was conducted using the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) from pieces of the dried specimens. ITS region was amplified using primers ITS1F and LR3/ITS4B [Citation8,Citation9]. Sequencing of the amplified PCR products of the specimens was performed using the services of Macrogen Inc. (Seoul, Korea). The reference sequences were obtained from the GenBank database (www.ncbi.nlm.nih.gov/genbank/). The obtained sequences were assembled, proofread, and edited using MEGA v. 6. [Citation10]. The sequences were aligned using MAFFT 7.130 [Citation11]. The best-fitting DNA substitution models for each dataset were tested with jModeltest 2.1.10 using the Bayesian information criterion (BIC) with default options [Citation12]. Bayesian (BI) analysis was performed using MrBayes 3.2.3 on XSEDE with a total of 20 million generations, with sampling every 1000 generations [Citation13]. We eliminated the first 25% of the sample trees as the burn-in and constructed a 50% majority rule consensus tree.
The maximum likelihood (ML) analyses were performed using RAxML with the GTR + G model of the evolution and 1000 bootstrap replicates [Citation14]. All analyses were carried out on the CIPRES web portal [Citation15]. The trees were edited in FigTree-version 1.4.3 [Citation16] and Adobe Illustrator CS6 (Adobe Systems, Inc., San Jose, CA, USA).
2.3. Morphological observation
Detailed macro- and microscopic morphological features of the species were investigated to confirm their identities. The color of the macroscopic structures was standardized following the Munsell colors [Citation17]. The observation of the microscopic features was performed from slide preparations mounted in 5% potassium hydroxide (KOH) under an Olympus BX51 light microscope (Tokyo, Japan) [Citation18]. At least 30 basidiospores and basidia structures were taken and measured. The following abbreviations are used in this paper: L = mean spore length, W = mean spore width, n = number of spores from a given specimen, Q = variation in the L/W ratios. In the case of the basidiospores, 5% of the measurements at each end of the range were given in parentheses.
3. Results
3.1. Molecular approach
The DNA sequences of the ITS region of the five mushrooms were obtained from Macrogen Inc. (Seoul, Korea). The species that most closely aligned with the ITS sequences were identified using a BLAST search via the NCBI database. The five mushrooms KUC20180712-50, KUC20180712-66, KUC20180712-65, KUC20180911-29, and KUC20180711-11 were identified as Calocybe decolorata (GenBank accession no. NR_156302) with 99% homology (676/681 bp), Crepidotus brunnescens (MF621031) with 97% homology (268/277 bp), Mycena pearsoniana (JN182200) with 99% homology (617/624 bp), Psathyrella phegophila (FN396129) with 99% homology (639/645 bp), and Psathyrella sulcatotuberculosa (KJ138423) with 98% homology (627/639 bp).
The five species were classified through phylogenetic analysis with the BI and ML method using the ITS sequence datasets. In the BI analysis of Calocybe decolorata KUC20180712-50 was assigned as HKY + G to the best-fit model, which contained 37 taxa and 871 nucleotide sequences; Crepidotus brunnescens KUC20180712-66 was assigned as GTR + G to the best-fit model, which contained 40 taxa and 792 nucleotide sequences; Mycena pearsoniana KUC20180712-65 was assigned as HKY + I+G to the best-fit model, which contained 24 taxa and 634 nucleotide sequences; P. phegophila KUC20180911-29 and Psathyrella sulcatotuberculosa KUC20180711-11 were assigned to the best-fit model as GTR + G by jModeltest 2.1 and contained 49 taxa and 794 nucleotide sequences. The ML trees showed the same topology as that obtained from the BI trees. In the phylogenetic tree, the C. decolorata KUC20180712-50 and C. brunnescens KUC20180712-66 loci were well separated from the other taxa and had high bootstrap values and posterior probabilities ( and ). M. pearsoniana KUC20180712-65 formed a monophyletic group with Mycena pearsoniana which showed a high bootstrap value and posterior probability (). The closest sister groups of P. phegophila and P. sulcatotuberculosa were Psathyrella subspadiceogrisea and P. singeri, respectively (). They also formed monophyletic groups with high bootstrap values and posterior probabilities.
Figure 2. Phylogenetic tree based on ITS region of Calocybe species using maximum likelihood (ML) analysis. Numbers at the nodes indicate Bayesian posterior probabilities (PP) > 0.75 and ML bootstrap proportion (BP) > 70% as PP/BP. A hyphen (“-”) indicates values PP < 0.75 or BP < 70%. The specimen and GenBank numbers are shown after the scientific name. The unrecorded Korean species is in bold letters within a red box. The scale bar indicates nucleotide substitutions per position.
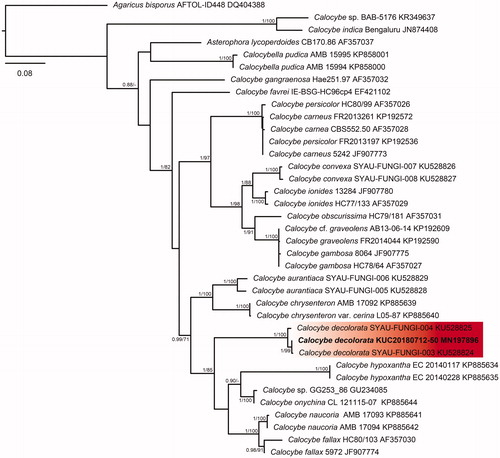
Figure 3. Phylogenetic tree based on ITS region of Crepidotus species using Maximum Likelihood (ML) analysis. Numbers at the nodes indicate Bayesian posterior probabilities (PP) > 0.75 and ML bootstrap proportion (BP) > 70% as PP/BP. A hyphen (“-”) indicates values PP < 0.75 or BP < 70%. The specimen and GenBank numbers are shown after the scientific name. The unrecorded Korean species is in bold letters within a red box. The scale bar indicates nucleotide substitutions per position.
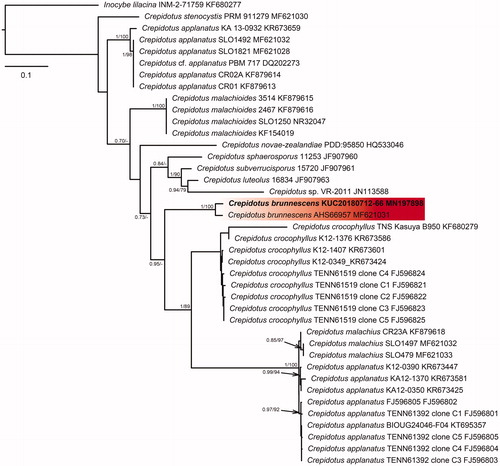
Figure 4. Phylogenetic tree based on ITS region of Mycena species using Maximum Likelihood (ML) analysis. Numbers at the nodes indicate Bayesian posterior probabilities (PP) > 0.75 and ML bootstrap proportion (BP) > 70% as PP/BP. A hyphen (“-”) indicates values PP < 0.75 or BP < 70%. The specimen and GenBank numbers are shown after the scientific name. The unrecorded Korean species is in bold letters within a red box. The scale bar indicates nucleotide substitutions per position.
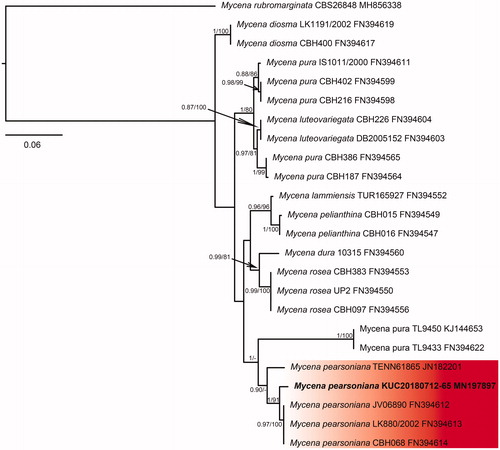
Figure 5. Phylogenetic tree based on ITS region of Psathyrella species using Maximum Likelihood (ML) analysis. Numbers at the nodes indicate Bayesian posterior probabilities (PP) > 0.75 and ML bootstrap proportion (BP) > 70% as PP/BP. A hyphen (“-”) indicates values PP < 0.75 or BP < 70%. The specimen and GenBank numbers are shown after the scientific name. The unrecorded Korean species is in bold letters within a red box. The scale bar indicates nucleotide substitutions per position.
3.2. Taxonomy
Calocybe decolorata X.D. Yu & J.J. Li, Mycologia 109(1): 61 (2017) ( and ).
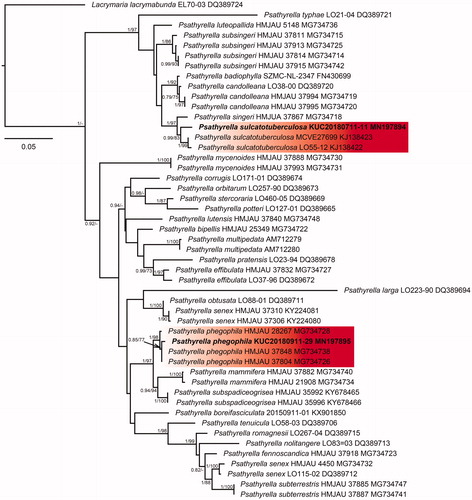
Figure 6. The basidiocarp surface (a) and (b) and microscopic features (c–e) of Calocybe decolorata KUC20180712-50. (c) Basidiospores; (d) Basidia; (e) Cystidia.
Pileus concave plane in age, 40 mm across; surface yellow (10YR 8/8) to dark yellowish-brown (10 YR 3/6) at the center, hygrophanous, margin uplifted and flexuous. Lamella crowded, narrow, decurrent, fragile, white. Stipe 40 × 3.5 mm, cylindrical, smooth, somewhat expanded at base, fibrillose, yellow to orange. Basidia clavate with 4 sterigmata (14.6–)15.1–20.9(–21.2) × 4.6–5.4(–6.8) µm. Basidiospores subglobose, 2.6–3.8(–3.9) × 2.4–3.1(–3.3) µm, L = 3.1 µm, W = 2.7 µm, Q =1.0–1.3(–1.4) (n = 30/1). Cystidia (19.1–)20.7–45.4(–58) × (3.6–)5.1–12(–12.5) µm, thin-walled, various shapes (subcylindrical to fusiform, ventricose with rounded ends, lageniform).
Specimen examined: Korea, Gyeongsangbuk-do, Cheongsong-gun, Juwang National Park, Mt. Juwang, 36°26′26.0″ N, 129°08′17.6″ E, July 12, 2018, Sun Lul Kwon, KUC20180712-50 (NIBRFG0000505334; GenBank accession no. MN197896), form solitary around leaf humus under mixed forest near Dalgi Falls.
Remark: Calocybe decolorata was recently reported as a new species, with the first record appearing in northeastern China. It grows gregarious on the soil in hardwood-conifer forests [Citation19]. The micro- and macroscopic structures of C. decolorata KUC20180712-50 were well matched with the original description. According to Li et al. [Citation19], the color of the gills of C. decolorata change to blue when bruised. However, the discoloration of the gills was not observed in the present study. Calocybe buxea has been reported to have discolored gills after bruising much like C. decolorata [Citation20]. However, C. buxea is distinguished by a white pileus and larger basidiocarp [Citation21].
Crepidotus brunnescens Hesler & A.H. Sm., North American species of Crepidotus: 52 (1965) ( and ).
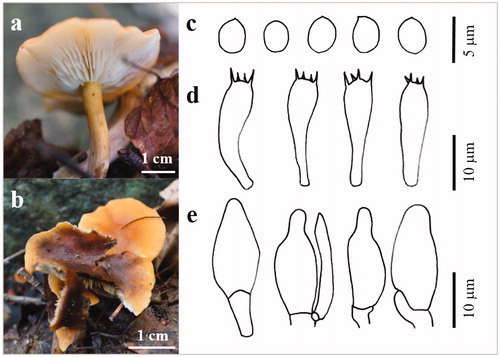
Figure 7. The basidiocarp surface (a) and (b) and microscopic features (c–e) of Crepidotus brunnescens KUC20180712-66. (c) Basidiospores; (d) Basidia; (e) Cystidia.
Pileus broadly attached, 8–12 mm across, sessilis, conchate to fan-shaped; surface white (2.5Y 8/1) to very pale brown (2.5Y 8/2), hygrophanous, striate to near base, thinly fibrillose to fibrillose-pubescent. Lamellae subclose to close, moderately broad, interspersed with narrow ones. Basidia clavate with 4 sterigmata (19.8–)19.9–26.5(–27.0) × (6.4–)6.5–8.3(–8.7) µm, thin-walled. Basidiospores globose to less ovoid, yellowish‐brown, wall thickened, 5.7–6.8(–7.0) × (4.5–)5.0–6.5 µm, L = 6.2 µm, W = 5.8 µm, Q = 1–1.2(–1.3) (n = 33/1), yellowish in KOH. Cheilocystidia and Pileocystidia morphologically similar, 25.9–53.8 × 4–8.7 µm, clavate, cylindric‐constricted, often subcapitate, ventricose, lageniform. Hyphae monomitic (7.1–)6.7 × 3.6(–3.1) µm. Clamp connection present.
Specimen examined: Korea, Gyeongsangbuk-do, Cheongsong-gun, Juwang National Park, Mt. Juwang, 36°26′26.0″ N, 129°08′17.6″ E, July 12, 2018, Sun Lul Kwon, KUC20180712-66 (NIBRFG0000505336; GenBank accession no. MN197898), form grouped on hardwood near Dalgi Falls.
Remark: Crepidotus brunnescens is a rare species that has been described only based on an original material collected in Michigan, USA [Citation22]. It produces fruiting bodies on hardwood logs and stumps in summer. C. brunnescens is a member of Crepidotus sect. Sphaerula, which is distinguished by sessile fruiting body or that with a pseudostipe. The color of pileus is pallid grayish to Prout’s brown and the surface of pileus is finely fibrillose to fibrillose-pubescent, according to the original description [Citation22]. The macro- and micromorphology of specimen KUC20180712-66 was well matched with the original description. C. brunnescens has a similar morphology to that of Crepidotus stenocystis. However, the latter is distinguished from the former by basidia size [Citation22,Citation23].
Mycena pearsoniana Dennis ex Singer, Sydowia 12 (1–6): 233 (1959) ( and ).
Figure 8. The basidiocarp surface (a) and (b) and microscopic features (c–e) of Mycena pearsoniana KUC20180712-65. (c) Basidiospores; (d) Basidia; (e) Cystidia.
Pileus plano-convex in age, 18 mm across; surface white (7.5Y 8/1) to light brown (7.5YR 6/4), translucent-striate, hygrophanous, paler margin. Lamella close, narrow, adnate to somewhat decurrent, dorsally intervenose with age. Stipe 64 × 2.1 mm, cylindrical, smooth, pruinose to puberulous above, glabrous. Basidia clavate with 4 sterigmata (16.8–)18.9–26.5 × 7.1–9.6(–10.4) µm, basidia clamp present. Basidiospores broadly ellipsoid, inamyloid, 6.5–9.1(–9.7) × 3.5–4.8 µm, L = 7.8 µm, W = 4.2 µm, Q = 1.6–2.1(–2.5) (n = 31/1). Cheilocystidia 24–43.8 × 10–18.2 µm, ventricose with rounded ends, lageniform. Pleurocystidia absent.
Specimen examined: Korea, Gyeongsangbuk-do, Cheongsong-gun, Juwang National Park, Mt. Juwang, 36°26′26.0″ N, 129°08′17.6″ E, July 12, 2018, Sun Lul Kwon, KUC20180712-65 (NIBRFG0000505335; GenBank accession no. MN197897), form solitary on soil around leaf humus under mixed forest near Dalgi Falls.
Remark: Mycena pearsoniana is a member of sect. Calodontes, which is known as the Mycena pura complex [Citation24]. It is distinguished morphologically from other species in sect. Calodontes by having inamyloid spores and by lacking pleurocystidia, according to the original description [Citation24]. It grows solitary or gregarious to cespitose on leaf humus, under deciduous trees or shrubs, and less frequently under conifers in summer to autumn. They are often found in moist habitats. This species has been reported in Denmark, Germany, Mexico, Norway, and the USA [Citation24–26]. According to Harder et al. [Citation26], M. pearsoniana does not have a consistent pattern of inamyloid and has a weak amyloid reaction. Although the collections from North American show weak amyloidity, the collections from North European TL-3966 show completely inamyloidity. In the case of Korean specimen, M. pearsoniana KUC20180712-65 appears inamyloid. Thus, the amyloid test was insufficient to be a reliable criterion for distinct M. pearsoniana. The basidiospore size of M. pearsoniana has been reported from American specimens (5.5–7.2 × 4–4.8 µm) [Citation24,Citation26] and European specimens (6–9 × 3.5–4.5 µm) [Citation27,Citation28]. In the case of Korean specimen, it is more comparable to the European collections.
Psathyrella phegophila Romagn., Persoonia Suppl 2: 282 (1985) ( and ).
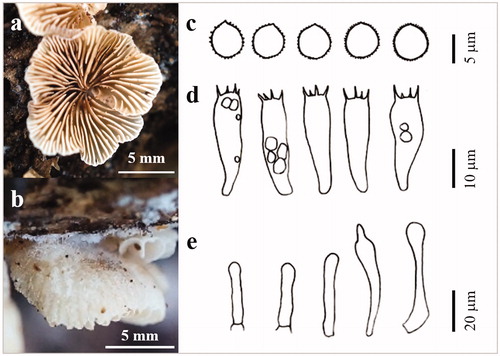
Figure 9. The basidiocarp surface (a) and (b) and microscopic features (c–e) of Psathyrella phegophila KUC20180912-29. (c) Basidiospores; (d) Basidia; (e) Cystidia.
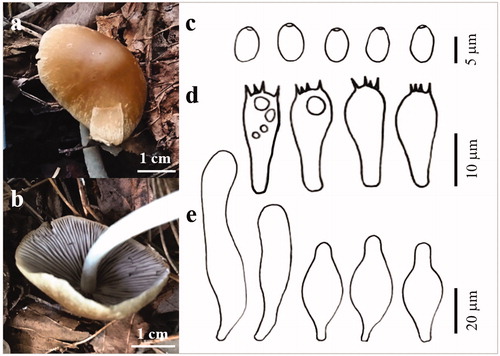
Pileus plano-convex in age, 32 mm across; surface reddish yellow (5YR 6/6), hygrophanous, paler margin. Lamellae close, narrow, adnate, even, purple-brown. Stipe 52 × 2–5 mm, cylindrical, fibrillose, rigid, white. Basidia clavate with 4 sterigmata (16.1–)17.8–22.4(–22.6) × (7–)8.1–9.7(–9.8) µm. Basidiospores ellipsoid, hyaline, smooth (6.3–)6.6–8.1(–8.6) × (3.7–)3.8–5.4(–5.7) µm, L = 7.4 µm, W = 4.7 µm, Q = 1.57 (n = 40/1). Cystidia 34.2–73.1× 11.9–18.9 µm, polymorph, utriform, lageniform, ventricose with rounded ends, clavate.
Specimen examined: Korea, Gyeongsangbuk-do, Cheongsong-gun, Juwang National Park, Mt. Juwang, 36°21′46.3″ N, 129°11′18.5″ E, September 11, 2018, Sun Lul Kwon, KUC20180911-29 (NIBRFG0000505333; GenBank accession no. MN197895), form solitary on leaf litter under mixed forest near Jusan Reservoir.
Remark: Psathyrella phegophila is a rare species [Citation29]. It is known to grow gregarious under beech (Fagus sylvatica) trees, on leaf litter, and soil rich in humus or on decaying wood [Citation29]. However, in this study, P. phegophila was found to be solitary on leaves. P. phegophila has been rarely reported in China, France, the United Kingdom, Switzerland, Spain, and Morocco [Citation29,Citation30]. P. phegophila is difficult to distinguish from Psathyrella fusca at the macroscopic level [Citation29]. At the microscopic level, the spore size of P. fusca is smaller than that of P. phegophila and the lamellae edge is very diverse. P. subspadiceogrisea is characterized by basidiospores that are rarely oval and a pileus without an umbo [Citation30]. Thus, they have separate morphological features.
Psathyrella sulcatotuberculosa (J. Favre) Einhell., Berichte der Bayerischen Botanischen Gasellschaft 47: 123 (1976) ( and ).
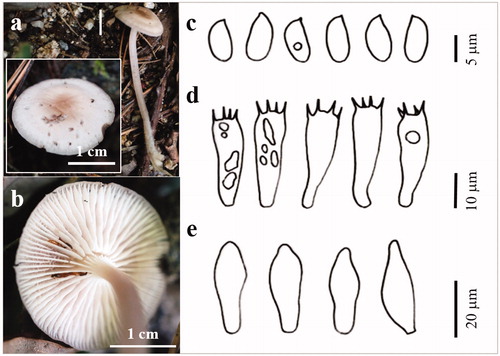
Figure 10. The basidiocarp surface (a) and (b) and microscopic features (c–e) of Psathyrella sulcatotuberculosa KUC20180711-11. (c) Basidiospores; (d) Basidia; (e) Cystidia.

Pileus fragile, convexo-expanded, 13 mm wide; surface white (7.5YR 9.5/1) to light yellowish-brown (10YR 6/4), glabrous, mat, hygrophanous, sulcate-tuberculose margin. Lamella subclose, adnate or adnexed, pale brown. Stipe 25 × 2.1 mm, cylindrical, straight or flexuous, fibrillose. Basidia clavate with 4 sterigmata (14.4–)14.7–20.4(–20.5) × 6.9–9.3(–9.7) µm. Basidiospores broadly ellipsoidal, ovoid, hyaline, smooth (6.1–)6.3–7.6 × (4.1–)4.2–5.4(–5.5) µm, L = 6.9 µm, W = 4.7 µm, Q = 1.3–1.7(–1.8) µm (n = 32/1). Cheilocystidia polymorph, numerous, utriform, lageniform, clavate, pyriform, oblong, ellipsoid. Pleurocystidia absence.
Specimen examined: Korea, Gyeongsangbuk-do, Cheongsong-gun, Juwang National Park, Mt. Juwang, 36°23′43.9″N, 129°09′15.0″E, July 11, 2018, Sun Lul Kwon, KUC20180711-11 (NIBRFG0000505332; GenBank accession no. MN197894), form solitary on leaf litter under mixed forest near Yongchu Falls.
Remarks: Psathyrella sulcatotuberculosa is a very rare species. It grows solitary or gregarious on leaf litter, soil, and vegetal debris. It has been rarely reported in Germany, Italy, Switzerland, and Spain [Citation31]. To the best of our knowledge, this is the first report of this species in Asia. P. sulcatotuberculosa is distinguished by small basidiomata, a sulcate-tuberculose pileus margin, a rudimentary veil, and the absence of pleurocystidia [Citation31]. P. sulcatotuberculosa KUC20180711-11 has the same microscopic structures as those in the original description. The color of the basidiocarp of KUC20180711-11 appeared to have been decolorized over time. The microscopic features of P. sulcatotuberculosa were similar to those of P. singeri. However, plano-umbonate and reddish purple-brown margin of the pileus of P. singeri were distinguished from P. sulcatotuberculosa [Citation31,Citation32]. P. sulcatotuberculosa is closely related to Psathyrella candolleana, which has been reported in Korea. However, the latter species is distinguished from the former by features of cheilocystidia, basidiospores brown in 5% KOH, and molecular data [Citation30,Citation32].
4. Discussion
In Korea, a total of 17 mountains were designated as national parks. Thus, biodiversity has been maintained despite the rapid development of Korea. However, knowledge about macrofungal diversity is still limited. Therefore, the National Institute of Biological Resources has started to investigate macrofungal diversity. In this study, five mushrooms that had not previously been recorded in Korea were identified using sequence-based identification. For the precise analysis of the candidates, phylogenetic analysis and morphological analysis were carried out. As a result, these fungi were identified as Calocybe decolorata, Crepidotus brunnescens, Mycena pearsoniana, P. phegophila, and P. sulcatotuberculosa.
Calocybe Kühner ex Donk are distributed worldwide, including in the neotropics, and approximately 87 species have been recorded globally [Citation20,Citation33]. However, only five Calocybe species had previously been recorded in Korea [Citation34]. The genus Calocybe is identified by the color of the pileus and the small basidiospores [Citation19]. Calocybe decolorata was reported recently in China [Citation19]. It is characterized by the change in the color of the gills when bruised, but no such color change was observed in the present study because the specimens were already bruised and seemed to be aged.
Crepidotus (Fries) Staude has been regarded as a member of Crepidotaceae based on traditional morphological studies [Citation35]. However, through phylogenetic analysis, the closest sister species of Crepidotus was identified as Inocybe (Fr.) Fr. [Citation36]. Thus, it now belongs to Inocybaceae, which has pleurotoid basidiomata. Crepidotus is characterized by the absence of clamp connections on the epicuticular hyphae of the basidiocarp [Citation22]. Despite the fact that 456 Crepidotus species have been recorded worldwide, only 18 Crepidotus species were listed in Korea [Citation33,Citation34].
The genus Mycena is characterized by a reddish-violaceous pileus, mostly amyloid spores, prominent voluminous cystidia and a raphanoid smell [Citation25]. The genus Mycena is also one of the largest genera recorded, with approximately 1853 species globally [Citation33] and 36 Mycena species have been recorded in Korea [Citation34]. Mycena pearsoniana belong to Mycena sect. Calodontes (Fr. ex Berk.) Quél. sensu MaasGeesteranus (Tricholomataceae s.l.). Nine Mycena species are in Mycena sect. Calodontes (Fr. Ex Berk.) Quél., including in two Mycena sect. Calodontes [Citation33,Citation34]. Because the species within the genus Mycena sect. Calodontes are morphologically similar to each other, they are difficult to identify [Citation27,Citation37]. Mycena sect. Calodontes has been morphologically distinguished by having inamyloid spores and by lacking pleurocystidia [Citation24]. According to Harder et al. [Citation25], this species is definitely not adjusted to the M. pearsoniana complex since some collections of M. pearsoniana show spores with weak amyloidity [Citation25], but the lack of pleurocystidia is still adjusted.
Psathyrella (Fr.) Quél. is distinguished by the fragile basidiomata, hygrophanous pileus, brown to black-brown spores, the presence of cheilocystidia, and the smooth or rarely granulose basidiospores [Citation30]. It is considered one of the most difficult macrofungi to study due to its fragile basidiomata. Although only 13 Psathyrella species have been recorded in Korea, the genus Psathyrella is one of the largest genera, of which 887 species have been reported globally [Citation31,Citation33,Citation34]. The P. sulcatotuberculosa collected in this study was found to be have a discolored pileus that was not observed in the original description.
The endangered species of Korea are managed by the National Institute of Biological Resources (https://www.nibr.go.kr/). To date, only one macrofungus has been recorded as an endangered species in Korea (i.e., Lampteromyces japonicus (current name = Omphalotus guepiniformis (Berk.) Neda)). Three species, C. brunnescens, P. phegophila, and P. sulcatotuberculosa, have been rarely found in Europe [Citation22,Citation29,Citation31]. Although they are not recorded in the IUCN Red List [Citation38], it is necessary to protect these species due to their rarity. The efforts to investigate macrofungi and conserve rare species are anticipated to enhance macrofungal diversity in Korea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hwang S-K, Kim J-H. Topographical landscapes and their controlling geological factors in the Juwangsan national park: welding facies and columnar joints. J Petrol Soc Korea. 2009;18:195–209.
- Ko P-Y, Hong K-S, Choe S-Y, et al. Distribution of spontaneously growing mushrooms in the Juwangsan National Park. J Mushrooms. 2018;16:65–69.
- Hibbett D, Abarenkov K, Kõljalg U, et al. Sequence-based classification and identification of Fungi. Mycologia. 2016;108(6):1049–1068.
- Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246.
- Kwon SL, Jang S, Kim M-J, et al. Identification of three wood decay fungi in Yeoninsan provincial park. Korea J Species Res. 2018;7:240–247.
- Cho HJ, Lee H, Park MS, et al. Macrolepiota in Korea: new records and a new species. Mycobiology. 2019;47(4):310–368.
- Tang L-P, Hao Y-J, Cai Q, et al. Morphological and molecular evidence for a new species of Rhodotus from tropical and subtropical Yunnan, China. Mycol Progress. 2014;13(1):45–53.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Darriba D, Taboada GL, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772–772.
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690.
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. IEEE Gateway Computing Environments Workshop (GCE); 2010 Nov 14; New Orleans, LA, USA. p. 1–8; 2010.
- Rambaut A. FigTree-version 1.4.3, a graphical viewer of phylogenetic trees. 2017 [cited 2016 Oct 4]. Available from: http://tree.bio.ed.ac.uk/software/figtree/
- Color BM. Munsell soil-color charts: with genuine Munsell® color chips. Grand Rapids (MI): Munsell Color; 2009.
- Largent D, Johnson D, Watling R. How to identify mushrooms to genus III: microscopic features. Eureka (CA): Mad River Press Inc.; 1977. p. 148.
- Li J-J, Wu S-Y, Yu X-D, et al. Three new species of Calocybe (Agaricales, Basidiomycota) from northeastern China are supported by morphological and molecular data. Mycologia. 2017;109(1):55–63.
- Singer R. The Agaricales in modern taxonomy. Koenigstein: Koeltz Scientific Books; 1986.
- Maire R. Études mycologiques (fascicule 2). Bull Soc Mycol France. 1930;XLVI:227.
- Hesler LR, Smith AH. North American species of Crepidotus. New York (NY): Hafner Publishing Company; 1965.
- Pouzar Z. Notes on some European species of the genus Crepidotus (Agaricales). Czech Mycol. 2005;57(3–4):299–305.
- Singer R. Fungí Mexicani, series secunda – Agaricales. Sydowia. 1958;12:221–243.
- Harder CB, Laessøe T, Kjøller R, et al. A comparison between ITS phylogenetic relationships and morphological species recognition within Mycena sect. Calodontes in Northern Europe. Mycol Progress. 2010;9(3):395–405.
- Harder CB, Lodge DJ, Petersen RH, et al. Amyloidity is not diagnostic for species in the Mycena pearsoniana complex (Mycena sectio Calodontes). Mycol Progress. 2012;11(3):725–732.
- Geesteranus RM. Some Myceneae of the Himalayan foothills. Persoonia. 1992;15:33–53.
- Aronsen A. A key to the Mycenas of Norway. [Internet]. 2011. Available from: www.mycena.no/
- Pérez-De-Gregorio MÀR, Carbó J. Dos Psathyrella interesantes halladas en Girona. Micol Veget Medit. 2010;25:23–32.
- Yan J-Q, Bau T. The Northeast Chinese species of Psathyrella (Agaricales, Psathyrellaceae. MycoKeys. 2018;33:85–102.
- Battistin E, Chiarello O, Vizzini A, et al. Morphological characterisation and phylogenetic placement of the very rare species Psathyrella sulcatotuberculosa. Sydowia. 2014;66:171–181.
- Smith AH. North American species of Psathyrella. Mem NY Bot Gard. 1972;24:1–633.
- Robert V, Stegehuis G, Stalpers J. The MycoBank engine and related databases. [Internet]. 2005. Available from: http://www.mycobank.org
- Lee Y, Lim Y, Kim J, et al. National list of species of Korea: Basidiomycota. Incheon: National Institute of Biological Resources; 2015.
- Consiglio G, Setti L, Robich G, et al. II Genere Crepidotus in Europa. Trento: Associazione Micologica Bresadola; 2008.
- Matheny PB. A phylogenetic classification of the Inocybaceae. McIlvainea. 2009;18:11–21.
- Rexer K-H. Die Gattung Mycena sl: Studien zu ihrer Anatomie, Morphologie und Systematik [PhD thesis]. Tübingen: Eberhard-Karls-Universität Tübingen; 1994.
- IUCN. The International Union for Conservation (IUCN) of Nature Red List of threatened Species. [Internet]. [Version 2019-3]. Available from: http://www.iucnredlist.org
