Abstract
Ganoderma lingzhi is a well-known source of natural fungal medicines which has been given for the treatment of several diseases. China is one of the major commercial producers of Ganoderma mushroom worldwide. However, with the expansion of the commercial cultivation, the occurrence of the fungal diseases on G. lingzhi has also been increased. The green mold disease symptoms were observed in the cultivation base of G. lingzhi in Zuojia Town, Jilin City, Jilin Province, China, causing the basidiomes to be rotten and withered, and the green mycelium layer generated gradually. The pathogenicity tests showed the same symptoms as appeared naturally in Zuojia mushroom base. Morphology characters revealed conidia green, ellipsoid, globose, 2.56–4.83 × 2.09–4.22 μm, length-width ratio was 1.1–1.2 (n = 10). Conidiophores trichoderma-like, often asymmetry, branches solitary, paired or in whorls of 3 phialides formed solitary, paired or in whorl, variable in shape, lageniform, sometimes ampulliform or subulate. While using molecular methodology, comparing with the sequences of Trichoderma hengshanicum from GenBank, the analyzed sequence showed 97.32% homology with the RPB2 sequences, 100% with the TEF1-α sequences. A fungus isolated from the diseased tissues was identified based on morphology and molecular studies as T. hengshanicum. This is the first report of T. hengshanicum causing the green mold disease of G. lingzhi in China.
Ganoderma lingzhi is a traditional Chinese medicinal mushroom [Citation1] Which is appreciated for anticancer effect as well as used to prevent all kind of diseases [Citation2,Citation3]. As a result, the commercially cultivated area and consumption of G. lingzhi is expanding annually [Citation4]. However, With the expansion of the cultivation of these mushrooms, the occurrence of fungal diseases on G. lingzhi has also been increased. Among the fungal diseases of G. lingzhi, green mold disease caused by Trichoderma spp. is the most devastating disease, and its incidence and severity have increased.
In 2018, we observed the basidiomes of G. lingzhi covered by a layer of green mycelium at G. lingzhi cultivation base in Zuojia Town, Jilin City, Jilin Province, China with incidence of over 7% area on cultivated G. lingzhi. In addition, the infected basidiomes were rotten and withered (). The diseased basidiomes were cut into 5 mm size pieces using the sterilized scalpel, and surface sterilization of infected tissues with 75% ethanol for 30 sec, washed twice with sterilized distilled water, dried with sterilized filter paper and placed into PDA plates and incubated at 25 °C for 5 days. later on, transferred to fresh PDA plates when the fungal hyphae emerged. The morphology, color, and other biological characters of the colony were observed on daily basis. After the incubation at 25 °C for 4 days, a small number of hyphae was picked with the inoculation needle and transferred onto a slide aseptically for morphological identification according to Yang [Citation5].
Figure 1. Green mold of G. lingzhi. (A) Deseased Basidiome of G. lingzhi covered with green mycelium layer founfed naturally; (B) About 14 days later, the whole peilus of G. lingzhi was covered with green mycelium; (C) Deseased basidiome of G. lingzhi under high temperature and humidity conditions.

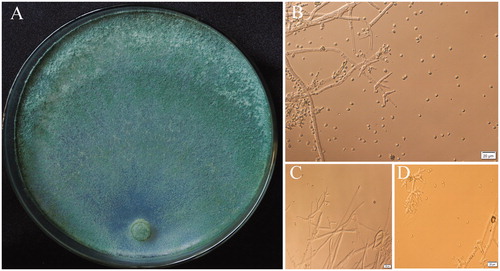
At the optimum growth temperature of 25 °C on PDA, the colony radius was 35 mm–38 mm after 3 days and the plate covered with mycelium after 5 days. The colony color was observed greenish, mycelium loose, aerial hyphae numerous, more numerous in the center of the colony (). Conidia starting after 2 days, formed on aerial hyphae near the colony center or in conidial pustules around the periphery of the colony. Conidia green, ellipsoid, globose, 2.56–4.83 × 2.09–4.22 μm, length-width ratio was 1.1–1.2 (n = 10). Conidiophores trichoderma-like, often asymmetry, branches solitary, paired or in whorls of 3. Phialides formed solitary, paired or in whorl, variable in shape, lageniform, sometimes ampulliform or subulate. No chlamydospores observed (). Distinct odor none, diffusing pigment absent. Morphological and cultural characteristics were accorded with T. hengshanicum [Citation6].
Figure 2. (A) The colony characteristics of T. hengshanicum, cultured on 2% PDA 5d; (B–D) Microscopic feature of T. hengshanicum Conidiophore, Phialides and Conidia.
Genomic DNA of the pathogen were extracted from the samples using the Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology Co., Hangzhou, China). Sequence translation elongation factor 1-alpha (TEF1-α) was amplified using primers EF1160R and EF595F [Citation7]. The second largest subunit of RNA polymerase II (RPB2) was amplified using primers fRPB2–5F and bRPB2–7R2 [Citation8]. The Polymerase chain reaction (PCR) mixture contained 10 μL of templat DNA, 25 μL of 2 × Es Taq MasterMix (Dye), 1 μL of each primer, and 13 μL of ddH2O, in a total volume of 50 μL. PCR parameters for the RPB2 and TEF1-α regions consisted of an initial 3 ̶ 5 min denaturation at 94 °C, 33 cycles at 94 °C 30 s, 53 °C ̶ 56°C for 30 s ̶ 40 s, 72 °C for 30 s ̶ 50 s and a final extension at 72 °C for 5 min, followed by immersion at 4 °C. Finally, gel electrophoresis was used to detect the product and sequences were completed by BGI Co., Ltd, Beijing, China. The TEF1-α and RPB2 sequences generated in this study were deposited in GenBank and compared with other sequences on GenBank using BLAST. According to the results, the closely matched sequences and related sequences described in were downloaded for phylogenetic analyses. The sequences were aligned with closely related strains by Clustal X 2.1 [Citation9]. The phylogenetic tree was constructed using the Maximum Likelihood (ML) method with 1000 bootstrap replicates by MEGA 7 (https://www.megasoftware.net/archived_version_active_download) [Citation10].
Table 1. Closely matched and related sequences downloaded and used for phylogenetic analyses in this study.
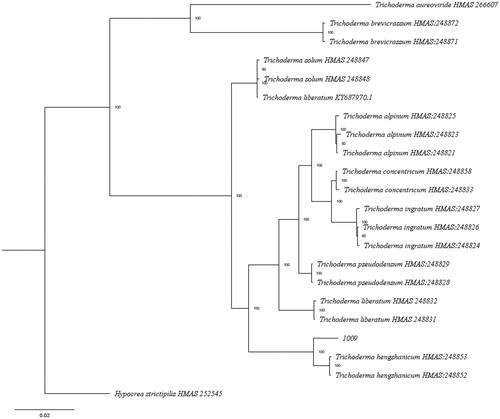
Comparing with the sequences of T. hengshanicum of GenBank, the analyzed sequence showed 97.32% homology with the RPB2 sequences (GenBank accession KY687992 and KY687991), 100% with the TEF1-α sequences (KY688055.1 and KY688054.1). TEF1-α and RPB2 sequences of 1009 and other 33 related taxa were used to perform ML analysis. The phylogenetic tree indicated that strain 1009 was related to T. hengshanicum (). Combining with morphological characteristics identification, strain 1009 was identified as T. hengshannicum.
Figure 3. The phylogenetic tree based on combined of TEF1-α and RPB2 sequences using Maximum Likelihood method with 1000 bootstrap replicates.
To check the pathogenicity of representative strain 1009, the experiments were carried out in duplicate. G. lingzhi HMJU10001 used to inoculate pathogen was stored at the microbiology laboratory of Jilin Agriculture Science and Technology College. The artificial inoculation was performed when the pilus of G. lingzhi grew to a large area with yellowish brown color and size 3–4 cm. All treatments were maintained at 25 °C to 30 °C and humidity 80% to 90% under controlled cultivation conditions. The pathogen block was inoculated with a spore suspension (1 × 107 conidia/mL). Different parts of the basidiome (peilus, mycelium tube, stipe) were used to inoculate with the help of syringe needles. The control block was inoculated with the suspension prepared from PDA alone and four repetitions per treatment were maintained while pathogenicity experiment was repeated twice. Changes in disease symptoms were observed and recorded every day for 14 days. Pathogens were reisolated from infected basidiomes at different disease stages (after every 2 days until 14 days of inoculation).
All of the inoculated basidiomes of G. lingzhi showed first symptoms after 24 h in which surface of the peilus, mycelium tube and stipe appeared taupe lesions. After 3 days, the peilus and stipe were mantled with white to green colonies and color became light, white villous mycelia grew from the inoculation site of mycelium tubes and aqua spores were produced around the site gradually. There were no obvious changes from macroscopic observation and never produced white villous mycelia also on the surface of peilus and stipe after 5 days, while brown rot was occurred on the diseased part of mycelium tubes and diseased range was diffused to 3.8-5.4 cm. With green colonies and abundant mycelia, the rotten range was greater than 1/2 of the whole G.lingzhi’s basidiome. Colonies of the diseased part of mycelium tubes were dark green and covered with a new layer of white spores again after the ninth day. After 14 days, the whole mycelium tubes of G. lingzhi were covered with green mycelia, while the basidiomes were rotten and withered (), however, the mushrooms grew up normally under the control block.
The pathogenicity experiments showed that the symptom of fruiting bodies inoculated with spore suspensions from the isolate was similar to that of the original symptomatic samples, Hence Koch’s postulates were proved. Based on the morphological and molecular characteristics and Koch’s postulates, we confirmed that the myco-pathogenic fungus T. hengshanicum was the causal agent of the disease on G. lingzhi. To our knowledge, this is the first report of Green Mold Disease caused by Trichoderma hengshanicum on Ganoderma lingzhi.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cao Y, Wu SH, Dai YC. Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Diversity. 2012;56:49–62.
- Su YZ, Sheen LY. An evidence-based perspective of Ganoderma Lucidum (Lucid Ganoderma) for cancer patients. In: Cho W, editor. Evidence-based anticancer materia medica. Dordrecht: Springer; 2011. p. 245–263.
- Lin ZP. The study annotated the discussion of Ganoderma Lucidum in “Shen Nong’s Herbal Classic” combinning Chinese and Western. Chin J Pharmacol Toxicol. 2019;33:645–646.
- Chyxx 2017. Analysis on supply demand and price trend of basswood Ganoderma lingzhi market in China 2017. Available from: https://www.chyxx.com/industry/201706/537394.html
- Yang HT. Classification and identification of trichoderma. Beijing: China Earth Press; 2009.
- Chen K, Zhuang WY. Discovery from a large-scaled survey of Trichoderma in soil of China. Sci Rep. 2017;7:37.
- Wendland J, Kothe E. Isolation of tef1 encoding translation elongation factor EF1a from the homobasidiomycete Schizophyllum commune. Mycol Res. 1997;101:798–802.
- Han YL, Wang D, Zhou J, et al. Intragenomic polymorphisms of the ITS, EF1-α and RPB2 sequences of Ganoderma austral. Mycosystema. 2019;38:679–685.
- Lin CY, Wei JD, Cheng HJ, et al. Embedded-based graphics processing unit cluster platform for multiple sequence alignments. Evol Bioinform Online. 2017;13:1176934317724764.
- Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321.
