Abstract
The family Peronosporaceae, an obligate biotrophic group of Oomycota, causes downy mildew disease on many cultivated and ornamental plants such as beet, cucumber, grape, onion, rose, spinach, and sunflower. To investigate the diversity of Peronosporaceae species in Korea, we performed morphological analysis for dried plant herbariums with downy mildew infections by two largest genera, Peronospora and Plasmopara. As a result, it was confirmed that there are five species of Peronospora and two species of Plasmopara, which have been so far unrecorded in Korea, as well as rarely known in the world; Pl. angustiterminalis (ex Xanthium strumarium), Pl. siegesbeckiae (ex Siegesbeckia glabrescens), P. chenopodii-ambrosioidis (ex Chenopodium ambrosioides), P. chenopodii-ficifolii (ex Chenopodium ficifolium), P. clinopodii (ex Clinopodium cf. vulgare), P. elsholtziae (ex Elsholtzia ciliata), and P. lathyrina (ex Lathyrus japonicus). In addition, their phylogenetic relationship was inferred by molecular sequence analysis of ITS, LSU rDNA, and cox2 mtDNA. By rediscovering the seven missing species and barcoding their DNA sequences, this study provides valuable insights into the diversity and evolutionary studies of downy mildew pathogens.
1. Introduction
The family Peronosporaceae is an obligate biotrophic pathogen group belonging to Oomycota. They cause downy mildew disease on a wide range of angiosperm plants [Citation1]. Notably, it led to severe damages on many economically cultivated and ornamental plants, including beet, broccoli, cucumber, grape, onion, rose, spinach, sunflower, and tobacco [Citation2]. The Peronospora is the largest genus in Peronosporaceae, including many notorious species, e.g. P. belbahrii (parasitic to sweet basil), P. destructor (onion), P. effusa (spinach), P. sparsa (rose), and P. tabacina (tobacco) [Citation1]. The genus Plasmopara is the second largest group in Peronosporaceae and includes a historically important species Plasmopara viticola, responsible for grape downy mildew [Citation3] that caused huge losses of grape production in Europe in the nineteen century [Citation4].
Morphological characteristics applied for modern taxonomy of oomycetes are often too variable to distinguish between close species of the Peronosporaceae. In addition, as they are unculturable in artificial media, cultural features are unavailable for classification and identification of this group. Instead, their high host specificity limited to a host genus or species is one of the most useful characters, although there are some exceptional species with a broad host range, e.g. Pseudoperonospora cubensis clade 1 [Citation5] and Plasmopara viticola clade 1 [Citation6,Citation7]. The recent advance of molecular phylogenetic analysis has led to marked changes and developments in taxonomy of two genera Peronospora and Plasmopara [Citation3,Citation8,Citation9]. However, long-forgotten species, which have been previously recorded but neither collected again or analyzed, are key pieces for solving the puzzle of studying their diversity and taxonomy. In some cases, however, it is still uncertain if such species are present or wrongly recorded by misidentification of pathogen or/and host plant. However, since most of them have been on unusual host plants, their inclusion is essential to understand the evolutionary relationship between pathogen and host plants. Unfortunately, they are mostly based on a single or few collection, which is frequently hard to access and even if accessed, it is challenging to obtain sequence data from such old collections.
During the surveys of the species diversity of Peronosporaceae in Korea [Citation10–14], we collected several rare species of Peronospora and Plasmopara, which have been previously recorded but not been subjected to modern taxonomic and phylogenetic approaches. The present study performed morphological and molecular phylogenetic analyses for the Korean specimens. Seven species of Peronospora and Plasmopara are recorded here; Pl. angustiterminalis (ex Xanthium strumarium), Pl. siegesbeckiae (ex Siegesbeckia glabrescens), P. chenopodii-ambrosioidis (ex Chenopodium ambrosioides), P. chenopodii-ficifolii (ex Chenopodium ficifolium), P. clinopodii (ex Clinopodium cf. vulgare), P. elsholtziae (ex Elsholtzia ciliata), and P. lathyrina (ex Lathyrus japonicus).
2. Materials and methods
2.1. Oomycete samples
Plants with oomycete infection were collected from different sites of Korea. Information on the dried herbarium samples selected for morphological and molecular phylogenetic analyses is provided in .
Table 1. Information of Oomycetes specimens used in this study.
2.2. Morphological analysis
Conidiophores (or Sporangiophores) and conidia (or sporangia) formed under the infected leaves were transferred to a drop of lactic acid on a slide glass, covered with a coverslip, and gently warmed up using an alcohol lamp. The microscope preparations were examined under a DIC-light microscope (BX53F, Olympus, Tokyo, Japan) and photographed with a DigiRetina 16 M digital camera (Tucsen, Fuzhou, China).
2.3. DNA extraction, PCR, and sequencing
Genomic DNA was extracted from the infected plant leaf using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The mitochondrial cytochrome c oxidase subunit 2 (cox2) gene was amplified with primers cox2-F [Citation15] and cox2-RC4 [Citation16]. An additional marker for Peronospora species, the internal transcribed spacer (ITS) region was amplified with primers ITS1-O [Citation17] and LR0 [Citation18], and for Plasmopara species the D1-3 region of the 28S ribosomal large subunit (LSU) was amplified with primers LR0R [Citation18] and LR6-O [Citation19]. PCR products were purified and sequenced by a DNA sequencing service (Macrogen Inc., Seoul, Korea), with the same primers used for amplification.
2.4. Phylogenetic analysis
Sequences of the ITS, LSU rDNA, and cox2 mtDNA were edited using the DNASTAR software package ver. 5.05 (DNAStar Inc., Madison, WI). The sequences of each locus were aligned using the Q-INS-I algorithm [Citation20] in MAFFT 7 [Citation21], along with the reference sequences retrieved from GenBank. Maximum likelihood (ML) and minimum evolution (ME) methods were used to infer the phylogenetic trees. For ML analysis, 1000 rounds of random addition of sequences as well as 1000 fast bootstrap replicates were performed using RAxML 7.0.3 [Citation22] with GTRCAT model. ME analysis was performed using MEGA 7.0 [Citation23] with the default settings of the program, except for replacement with the Tamura-Nei model, and the robustness of the ME tree was evaluated with 1000 bootstrap replicates.
3. Results and discussion
Phylogenetic relationships of two Plasmopara species parasitic to Xanthium strumarium and Siegesbeckia glabrescens were inferred using ML and ME analyses of LSU rDNA and cox2 mtDNA sequences. The reference sequences of other Plasmopara species were retrieved from a phylogenetic analysis of Choi et al. [Citation24], and their GenBank accession numbers were shown in . As two topologies generated from ML and ME inferences were completely compatible (data not shown), except for minor differences in supporting values, only the ML tree is shown for each locus ( for LSU and for cox2), with ML and ME bootstrap values higher than 60% at the first and second position above the branches. In LSU and cox2 trees, it was confirmed that Plasmopara specimen ex X. strumarium matches with a reference sequence of Pl. angustiterminalis in Bulgaria and that Pl. siegesbeckiae ex S. glabrescens is distinguished between other Plasmopara species.
Figure 1. Maximum likelihood trees of Plasmopara species based on the LSU ribosomal DNA (D1/D2/D3) sequences (A) and the cox2 mitochondrial DNA sequences (B), with support values in minimum evolution inference. Bootstraping support values (minimum evolution/maximum likelihood) higher than 60% are given above the branches. The scale bar equals the number of nucleotide substitutions per site.
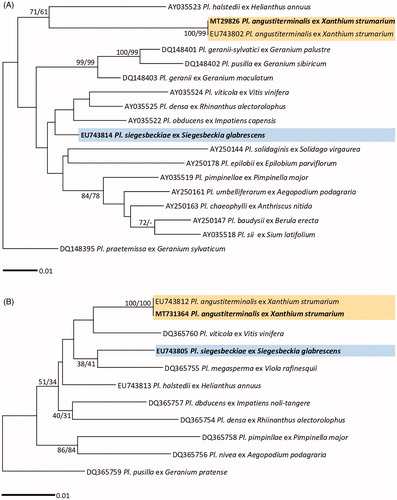
Table 2. Genbank accession numbers of the ITS, 28S rDNA, and cox2 mtDNA sequences used for the phylogenetic study.
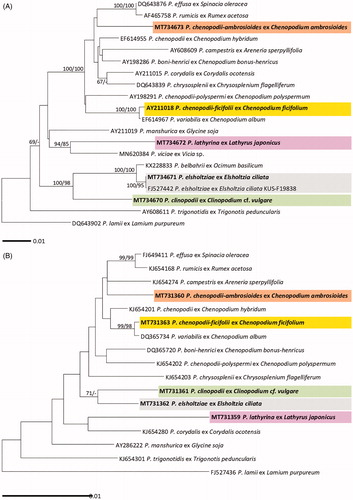
For Peronospora species ML and ME trees were constructed using ITS rDNA and cox2 mtDNA sequences ( for ITS and for cox2). Five Peronospora specimens in Korea, parasitic on Chenopodium ambrosioides, Chenopodium ficifolium, Clinopodium cf. vulgare, Elsholtzia ciliata, and Lathyrus japonicus, were compared with the reference sequences of other Peronospora species which were investigated in previous studies [Citation9,Citation12]. As this study initially provides the sequence data for these species, except for P. elsholtziae, there was no sequence available in GenBank. The GenBank accession numbers of the sequences used in this study were shown in . In both ITS and cox2 trees, P. chenopodii-ficifolii ex C. ficifolium was closely related to P. variabilis ex C. album, with the maxim supporting value in ITS tree and high values of 99/98 in cox2 tree, but other four species of Peronospora, P. chenopodii-ambrosioidis (ex Chenopodium ambrosioides), P. clinopodii (ex Clinopodium cf. vulgare), P. elsholtziae (ex Elsholtzia ciliata), and P. lathyrina (ex Lathyrus japonicus), formed each independent branch. In ITS tree, P. lathyrina was clearly distinguished from P. viciae, another downy mildew species on Fabaceae.
Figure 2. Maximum likelihood trees of Peronospora species based on the internal transcribed spacer (ITS) sequences (A) and the cox2 mitochondrial DNA sequences (B), with support values in minimum evolution inference. Bootstraping support values (minimum evolution/maximum likelihood) higher than 60% are given above the branches. The scale bar equals the number of nucleotide substitutions per site.
Through a microscopic examination, the morphological characteristics of two Plasmopara and five Peronospora species were described in detail below. Each species resembles ones of the original description. Based on phylogenetic and morphological analyses, it was confirmed that there are five species of Peronospora (P. chenopodii-ambrosioidis, P. chenopodii-ficifolii, P. clinopodii, P. elsholtziae, and P. lathyrina) and two species of Plasmopara (Pl. angustiterminalis and Pl. siegesbeckiae), all of which have been unrecorded in Korea.
4. Taxonomy
Plasmopara angustiterminalis Novot., Bot. Zh. SSSR 47(7): 979 (1962) [MB#337101] ().
Figure 3. Morphological characteristics of two Plasmopara species, Pl. angustiterminalis ex Xanthium strumarium (A–E) and Pl. siegesbeckiae ex Siegesbeckia glabrescens (F–J). (A & F), Sporangiophores; (B, C, G, H), Ultimate branchlets; (D, E, I, J), Sporangia (scale bars: 100 µm for sporangiophores, 10 µm for ultimate branchlets, and 10 µm for sporangia).
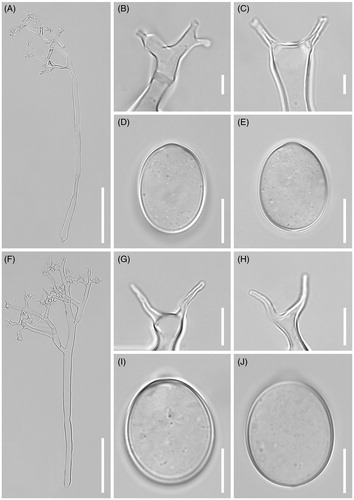
Description: Lesions commonly present on leaves, causing discoloration of the tissues and deformation of the attacked organs, polyangular, vein-limited, frequently covering larger areas by coalescing; infected tissues become necrotic. Down hypophyllous, whitish, consisting of agglomerated to scattered sporangiophores, felt-like, dense. Haustoria intracellular, not branched, flask-shaped vesicle, symmetrical, 11–20 μm diam., with a stalk at the part of entry into the host cell, surrounded by sheaths with 0.5–2 µm thickness. Sporangiophores emerging through stomata, subhyaline, straight to substraight, (200-)250-400(-550) µm long (n = 50); trunk straight, (150-)200-250(-400) µm long, 6–10 µm wide, base not or somewhat swollen, up to 11 µm wide, callose plugs commonly present; branches arising at a right angle to the main axis, monopodial, of 3–5 orders; callose plugs frequently present. Ultimate branchlets in pairs or three, straight, 4–14 µm long, 1–2.5 µm wide at the base; tip truncate or cup-like, rarely swollen. Sporangia subhyaline, broadly ellipsoidal, but sometimes ellipsoidal, (17-)19-22(-24) (av. 19.9) µm long, (11-)13-15(-17) (av. 14.6) µm wide, l/w ratio 1.2-1.5 (av. 1.37, n = 50). Resting organs not seen.
Habitat: on living leaves of Xanthium strumarium.
Specimen examined: Korea, Gangwon-do, Jeongseon-gun, 37°15’29 "N 128°21'35"E, 7 September 2009, H. D. Shin & Y. J. Choi, NIBRFG0000503192 (KUS-F24490).
Typus: on Xanthium strumarium; Ukraine, Zakarpatska, July 1960, M. K. Chochrjakov.
Note: The morphological features of the Korean specimen are consistent with the original description of Pl. angustiterminalis [Citation25], except for the size of sporangia; in the Korean sample (av. 19.9 × 14.6 µm) it is smaller than original one (av. 25 × 16 µm), but close to its measurements of China (av. 19.39 × 15.0 µm) [Citation26] and Austria (ca. 12–27 × 9–18 µm) [Citation27]. Also, Komjáti et al. [Citation28] recorded this species displays a high variation in sporangia, as av. 41.1 ± 7.8 × 27.0 ± 4.6 µm. In BLASTn search of LSU and cox2 sequences, this Korea specimen was identical to a Hungarian sample of Pl. angustiterminalis (SOMF07198). This species seems common in Europe, but rare in other continents [Citation29]. In Asia, this species has been previously recorded in China [Citation26], and this is the first report of Pl. angustiterminalis in Korea.
Plasmopara siegesbeckiae (Lagerh.) J. F. Tao, Acta Mycol. Sin. 6 (2): 69 (1987) [MB#131964] ().
Description: Lesions commonly present on leaves, causing discoloration of the tissues and deformation of the attacked organs, poly-angular, vein-limited, frequently covering larger areas by coalescing; infected tissues become necrotic. Down hypophyllous, whitish, consisting of agglomerated to scattered sporangiophores, felt-like, dense. Haustoria intracellular, not branched, flask-shaped vesicle, symmetrical, 11–20 μm diam., with a stalk at the part of entry into the host cell, surrounded by sheaths with 0.5–2 µm thickness. Sporangiophores emerging through stomata, subhyaline, straight to substraight, (150-)200-350(-550) µm long (n = 50); trunk straight, (100-)150-250(-400) µm long, 6–10 µm wide, base not or somewhat swollen, up to 11 µm wide, callose plugs commonly present; branches arising at a right angle to the main axis, monopodial, of 3–5 orders; callose plugs frequently present. Ultimate branchlets in pairs or three, straight, 4–13 µm long, 1–2.5 µm wide at the base; tip truncate or cup-like, rarely swollen. Sporangia subhyaline, broadly ellipsoidal, but sometimes ellipsoidal, (17.1-)18-22(-24) (av. 19.9) µm long, (11-)13-16(-19) (av. 14.6) µm wide, l/w ratio 1.2-1.5 (av. 1.37, n = 100). Resting organs not seen.
Habitat: on living leaves of Siegesbeckia glabrescens.
Specimen examined: Korea, Gangwon-do, Hoengseong-gun, 37°32'20"N 128°6'40"E, 4 September 2005, H. D. Shin & Y. J. Choi, NIBRFG0000503193 (KUS-F21312).
Typus: on Siegesbeckia orientalis; China, July 1979, J. F. Tao (Herb. No. 1373).
Note: This species was first reported as Peronospora leptosperma var. siegesbeckiae by Lagerh in 1895. Since then, Tao and Qin [Citation30] have combined it under the genus Plasmopara and promoted to the specie level, Plasmopara siegesbeckiae. The morphological characteristics of the Korean specimen resemble the original description [Citation30], except for the narrower sporangia (11–19 versus 16.6–20.6 µm) and two different host species (Siegesbeckia glabrescens versus S. orientalis). Pl. siegesbeckiae has been recorded previously in China and presently in Korea, hinting at it as an East Asian species.
Peronospora chenopodii-ambrosioidis Golenia, Monogr. bot., Soc. bot. Polon. 13: 147 (1962) [MB#335900] ().
Figure 4. Morphological characteristics of five Peronospora species, P. chenopodii-ambrosioidis ex Chenopodium ambrosioides (A, F, K, P), P. chenopodii-ficifolii ex Chenopodium ficifolium (B, G, L, Q), P. clinopodii ex Clinopodium cf. vulgare (C, H, M, R), P. elsholtziae ex Elsholtzia ciliata (D, I, N, S), and P. lathyirna ex Lathyrus japonicus (E, J, O, T). (A–E), Conidiophores; (F–J), Ultimate branchlets; (K–T), Conidia (scale bars: 100 µm for conidiophores, 10 µm for ultimate branchlets, and 20 µm for conidia).
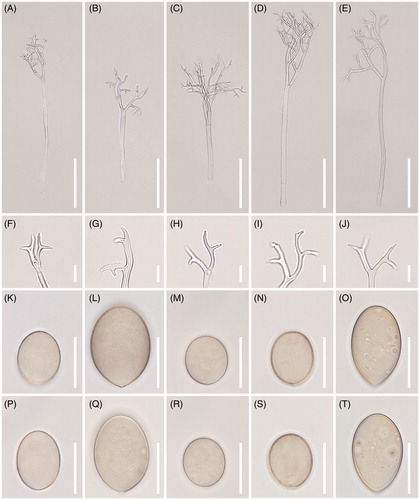
Description: At initial infection, lesions began yellowish then darken to become purplish, vein-delimited, resulting in a polyangular, mosaic appearance on leaves. Down present on the lower leaf surface of leaves, sparse to dense, felt-like, purplish, or beige. Conidiophores 220–500 µm long, colorless, straight; trunk 150–300 µm long, 5–10 µm wide, basal end slightly bulbous, up to 13 µm wide, callose plugs absent. Branches sub-dichotomously or monopodially branched 4–6 orders, substraight to slightly curved; ultimate branchlets flexuous to curved, 5–11 µm long, 1.5–2 µm wide, apex obtuse. Conidia olivaceous with gray tint, broadly ellipsoidal to ellipsoidal, greatest width median in most conidia, (17-)18-23(-28) (av. 21.5) µm long, (13-)15-19(-22) (av. 17.1) µm wide, l/w ratio = 1.2–1.5 (m = 1.32), pedicel absent in most conidia, sometimes appeared protruding. Resting organs not seen.
Habitat: on living leaves of Chenopodium ambrosioides.
Specimen examined: Korea, Jeju-do, 33°27'34"N 126°33'50"E, 12 November 2003, H. D. Shin & Y. J. Choi, NIBRFG0000503198 (KUS-F20063).
Typus: on Chenopodium ambrosioides; Poland, Poznań, no date information, A. Golenia (Herb. Inst. For Medicinal Plants Research, Poznań).
Note: This is the first report of P. chenopodii-ambrosioidis in Asia. All morphological features of the present specimen are consistent with the original description of Golenia [Citation31]; the size of conidia (av. 21.5 × 17.1 versus 22.5 × 17.2 µm), a length to width ratio of conidia (av. 1.32 versus 1.3), and the length of conidiophores (220–500 versus 192–640 µm). Although the downy mildew infections on C. ambrosioides have been recorded worldwide [Citation29], the causal agents have been often mis-identified as either P. farinosa or P. chenopodii, due to the complex nomenclatural history for Peronospora species infecting Chenopodiaceae plants [Citation12,Citation32,Citation33].
Peronospora chenopodii-ficifolii Sawada, Taiwan, Sotokufu Noji Shikenjo (Agr. Exp. Stat. Formosa): 9 (1916) [MB#176653] ().
Description: At initial infection, lesions began yellowish then darken to become purplish, vein-delimited, resulting in a polyangular, mosaic appearance on leaves. Down present on the lower leaf surface of leaves, sparse to dense, felt-like, purplish, or beige. Conidiophores (200-)250-350(-500) µm long, straight; trunk (100-)150-200(-300) µm long, 8–10 µm wide, basal end not differentiated, rarely bulbous, up to 15 µm wide. Branches sub-dichotomously branched 5–7 orders slightly curved to sigmoid; ultimate branchlets flexuous to curved, 6–20 µm long, apex obtuse. Conidia olivaceous to brown, broadly ellipsoidal to ellipsoidal, greatest width median in most conidia, (20-)26-35 µm long, 20–25 µm wide, l/w ratio = 1.2–1.5, pedicel present. Resting organs not seen.
Habitat: on living leaves of Chenopodium ficifolium.
Specimen examined: Korea, Namyangju-si, 37°54'44"N 127°2'0"E, 23 May 2016, H. D. Shin & Y. J. Choi, NIBRFG0000503199 (KUS-F29169).
Typus: on Chenopodium ficifolium; China (Taiwan), June 1916, no details about the collection were provided.
Note: All morphological characteristics fit well with the original description of P. chenopodii-ficifolii [Citation34]. After the first description, this species has been long forgotten due to the same reason with P. chenopodii-ambrosioidis. The Korean specimens have been previously classified under the name P. farinosa [Citation10] that has been recently rejected [Citation33]. In the present morphological and phylogenetic analyses, P. chenopodii-ficifolii is distinguishable from other species of Peronospora parasitic to Chenopodium spp., and thus we treat it as a distinct species. This species seems common in Europe, as well as East Asia, including China, Japan, and Korea [Citation29].
Peronospora clinopodii Terui, Ann. Phytopath. Soc. Japan: 524 (1978) [MB#319385] ().
Description: At initial infection, lesions began yellowish or light greenish, and then darken to become purplish, vein-delimited, resulting in a polyangular, mosaic appearance on leaves. Down present on the lower leaf surface of leaves, sparse to dense, felt-like, purplish or beige. Conidiophores protruding from stomata on the underside of leaves, erect, hyaline, sub-dichotomously to monopodially or dichotomously branched 5–8 orders, (150-)200-300(-400) µm long, 9–11 µm wide; basal end slightly swollen (av. 13 µm). Ultimate branchlets slightly curved, with somewhat different lengths, 5–10 µm. Conidia subglobose to broadly ellipsoidal, olivaceous to light brownish, (17-)19-23(-25) µm long (av. 20.4), (15-)17-20(-23) (av. 18.7) µm wide, with a length to width ratio of 1.1–1.25 (av. 1.15, n = 50); pedicel absent or appeared as a scar.
Habitat: on living leaves of Clinopodium cf. vulgare.
Specimen examined: Korea, Yangpyeong-gun, 37°30'10"N 127°43'24"E, 4 November 2003, H. D. Shin & Y. J. Choi, NIBRFG0000503197 (KUS-F20032).
Typus: on Clinopodium sachalinense; Japan, Aomori Pref., Mt. Ajara, 10 October 1974, M. Teuri
Note: Peronospora clinopodii was first reported by Terui [Citation35] in Japan. The morphological features of the present specimen are similar to the original description, except for the length of conidiophores (150–400 µm versus 300–500 µm). In addition, it was parasitic on Clinopodium cf. vulgare in Korea, but on Clinopodium sachalinense in Japan. P. clinopodii seems to distribute only in East Asia, including Korea and Japan.
Peronospora elsholtziae T. R. Liu & C. K. Pai, Acta Mycol. Sin. 4(1): 5 (1985) [MB#105621] ().
Description: At initial infection, lesions began yellowish then darken to become purplish, vein-delimited, resulting in a polyangular, mosaic appearance on leaves. Down present on the lower leaf surface of leaves, sparse to dense, felt-like, purplish, or beige. Conidiophores protruding from stomata on the underside of leaves, erect, hyaline, monopodially to rarely subdichotomously branched 5–7 orders, (300-)350-500(-600) µm long, 8–12 µm wide; basal end not or slightly swollen at the base up to 13 µm. Ultimate branchlets straight to curved, mostly in pair, with somewhat different lengths, 5–10 µm. Conidia broadly ellipsoidal to ellipsoidal, olivaceous to light brownish, (18-)20-22.5(-24) µm long (av. 20.9), (16-)18-20(-21) µm wide (av. 18.4), with a length to width ratio of 1.05–1.25 (av. 1.13, n = 50). Resting organs not seen.
Habitat: on living leaves of Elsholtzia ciliata.
Specimen examined: Korea, Chuncheon-si, Sporangia, 37°56'8"N 127°39'35"E, 1 June 2004, H. D. Shin & Y. J. Choi, NIBRFG0000503196 (KUS-F20252).
Typus: on Elsholtzia ciliate; China, Heilongjiang, Mi-Shan Reg., June 1981, T. R. Liu.
Note: Before the present study, P. elsholtziae has been recorded only in China [Citation29,Citation36], and this is the first record of P. elsholtziae in Korea. The morphological features of the present specimen are consistent with the original description [Citation36], although the conidia of the Korean sample (av. 20.9 × 18.4 µm) are somewhat smaller than the original ones (av. 23.9 × 20.8 µm).
Peronospora lathyrina Vienn.-Bourg., Bull. trimest. Soc. mycol. Fr. 66: 64 (1950) [MB#302539] ().
Description: Lesions commonly present on leaves, causing discoloration of the tissues and deformation of the attacked organs, poly-angular, vein-limited, frequently covering larger areas by coalescing; infected tissues become necrotic. Down hypophyllous, whitish, consisting of agglomerated to scattered conidiophores, felt-like, dense. Haustoria intracellular, not branched, flask-shaped vesicle, symmetrical, 11–20 μm diam., with a stalk at the part of entry into the host cell, surrounded by sheaths with 0.5–2 µm thickness. Conidiophores emerging through stomata, subhyaline, straight to substraight, (200-)250-400(-550) µm long (n = 50); trunk straight, (150-)200-250(-400) µm long, 6–10 µm wide, base not or somewhat swollen, up to 11 µm wide, callose plugs commonly present; branches arising at a right angle to the main axis, monopodial, of 3–5 orders; callose plugs frequently present. Ultimate branchlets in pairs or three, straight, 4–13 µm long, 1–2.5 µm wide at the base; tip truncate or cup-like, rarely swollen. Conidia subhyaline, broadly ellipsoidal, but sometimes ellipsoidal, (20-)22-26(-30) µm long (av. 23.6), (11-)13-15(-17) µm wide (av. 14.5), l/w ratio (1.2-)1.3-1.6(-1.7) (av. 1.55 n = 100). Resting organs not seen.
Habitat: on living leaves of Lathyrus japonicus.
Specimen examined: Korea, Samcheok-si, Miro-myeon, 37°24'52"N 129°6'2"E, 8 May 2003, H. D. Shin & Y. J. Choi, NIBRFG0000503195 (KUS-F19431).
Typus: on Lathyrus japonica; France, Alpes Mts, Pelvoux Mt., Gironde valley, between the village Vollouise and Glacier Blanc shelter, 22 August 1948, G. Viennot-Bourgin.
Note: Peronospora lathyrina was first described by Viennot-Bourgin [Citation37] in France. The conidial size and l/w ratio of the Korean sample are slightly larger than ones of the original description; av. 21.4 × 16.3 µm versus 23.6 × 14.5 µm in size and av. 1.31 versus 1.55 in l/w ratio. In addition, the host species of the Korean sample is different from the original specimen; Lathyrus japonicus versus Lathyrus latifolius. Taxonomy and nomenclature of Fabaceae-infecting downy mildews, including P. lathyrina, are still in confusion [Citation38], and further study using additional specimens from different host plants is essential to their correct species definition and identification.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Thines M, Choi YJ. Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology. 2016;106(1):6–18.
- Thines M, Telle S, Choi YJ, et al. Baobabopsis, a new genus of graminicolous downy mildews from tropical Australia, with an updated key to the genera of downy mildews. IMA Fungus. 2015;6(2):483–491.
- Schröder S, Telle S, Nick P, et al. Cryptic diversity of Plasmopara viticola (Oomycota, Peronosporaceae) in North America. Org Divers Evol. 2011;11(1):3–7.
- Gessler C, Pertot I, Perazzolli M. Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol Mediterr. 2011;50:3–44.
- Runge F, Choi YJ, Thines M. Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur J Plant Pathol. 2011;129(2):135–146.
- Rouxel M, Mestre P, Comont G, et al. Phylogenetic and experimental evidence for host-specialized cryptic species in a biotrophic oomycete . New Phytol. 2013;197(1):251–263.
- Rouxel M, Mestre P, Baudoin A, et al. Geographic distribution of cryptic species of Plasmopara viticola causing downy mildew on wild and cultivated grape in eastern North America. Phytopathology. 2014;104(7):692–701.
- Spring O, Voglmayr H, Riethmüller A, et al. Characterization of a Plasmopara isolate from Helianthus × laetiflorus based on cross infection, morphological, fatty acids and molecular phylogenetic data. Mycol Progress. 2003;2(3):163–170.
- Göker M, Voglmayr H, Riethmüller A, et al. How do obligate parasites evolve? A multi-gene phylogenetic analysis of downy mildews. Fungal Genet Biol. 2007;44(2):105–122.
- Shin HD, Choi YJ. Peronosporaceae of Korea. Suwon: National Institute of Agricultural Science and Technology; 2006.
- Choi YJ, Constantinescu O, Shin HD. A new downy-mildew of the Rosaceae: Peronospora oblatispora sp. nov. (Chromista, Peronosporales). Nova Hedw. 2007;85(1):93–101.
- Choi YJ, Denchev CM, Shin HD. Morphological and molecular analyses support the existence of host-specific Peronospora species infecting Chenopodium. Mycopathologia. 2008;165(3):155–164.
- Choi YJ, Shin HD, Voglmayr H. Reclassification of two Peronospora species parasitic on Draba in Hyaloperonospora based on morphological and molecular phylogenetic data. Mycopathologia. 2011;171(2):151–159.
- Choi YJ, Hong SB, Shin HD. Diversity of the Hyaloperonospora parasitica complex from core brassicaceous hosts based on ITS rDNA sequences. Mycol Res. 2003;107(11):1314–1322.
- Hudspeth DSS, Nadler SA, Hudspeth MA. COX2 molecular phylogeny of the Peronosporomycetes. Mycologia. 2000;92(4):674–684.
- Choi YJ, Beakes G, Glockling S, et al. Towards a universal barcode of oomycetes-a comparison of the cox1 and cox2 loci. Mol Ecol Resour. 2015;15(6):1275–1288.
- Bachofer M. Molekularbiologische Populationsstudien an Plasmopara halstedii, dem Falschen Mehltau der Sonnenblume. Stuttgart: University of Hohenheim; 2004.
- Moncalvo J-M, Wang H-H, Hseu R-S. Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia. 1995;87(2):223–238.
- Riethmuller A, Voglmayr H, Goker M, et al. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia. 2002;94(5):834–849.
- Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Choi YJ, Kiss L, Vajna L, et al. Characterization of a Plasmopara species on Ambrosia artemisiifolia, and notes on P. halstedii, based on morphology and multiple gene phylogenies. Mycol Res. 2009;113(10):1127–1136.
- Novotel’nova NS. Plasmopara halstedii (Farl.) Berl. et De Toni kak sbornyj vid (Obosnovanie k taksonomicheskomu podrazdeleniyu roda Plamopara na slozhnotsvetnykh). Bot Zhurn. 1962;47:970–981.
- Tao J. Plasmopara. In: Yu Y, editor. Flora fungorum sinicorum. Vol. 6. Peronosporales. Beijing: Science Press; 1998. p. 329–368.
- Bedlan G, Thines M. Erstnachweis von Plasmopara angustiterminalis Novot. an Xanthium strumarium in Österreich. Linzer Biol Beitr. 2009;41:1631–1633.
- Komjáti H, Walcz I, Virányi F, et al. Characteristics of a Plasmopara angustiterminalis isolate from Xanthium strumarium. Eur J Plant Pathol. 2007;119(4):421–428.
- Farr DF, Rossman AY. Fungal Databases. U.S. National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/. 2019.
- Tao JF, Qin Y. Taxonomic studies on the genus Plasmopara of China III. New species, new combination and new record of Plasmopara on family Compositae. Acta Mycol Sin. 1987;6:65–73.
- Golenia A. Badania nad grzybem Peronospora ambrosioides sp. n. I pokrewnymi wroslikami. Monogr Bot. 1962;13:121–177.
- Choi YJ, Klosterman SJ, Kummer V, et al. Multi-locus tree and species tree approaches toward resolving a complex clade of downy mildews (Straminipila, Oomycota), including pathogens of beet and spinach. Mol Phylogenet Evol. 2015;86:24–34.
- Choi YJ, Thines M. (2288) Proposal to reject the name Botrytis farinosa (Peronospora farinosa) (Peronosporaceae: Oomycetes). Taxon. 2014;63(3):675–676.
- Tanaka T. New Japanese Fungi Notes and Translations—VI. Mycologia. 1919;11(2):80–86.
- Terui M. New or interesting downy midews found in Mt. Ajara, Aomori, Japan. Jpn J Phytopathol. 1978;44(4):523–524.
- Liu TR, Pai CK. Some new species Peronosporaceae in China. Mycosystema. 1985;4:5–11.
- Viennot-Bourgin G. Mycological notes. Bull Soc Myco Fr. 1950;66:58–70.
- García-Blázquez G, Göker M, Voglmayr H, et al. Phylogeny of Peronospora, parasitic on Fabaceae, based on ITS sequences. Mycol Res. 2008;112(5):502–512.
