Abstract
Three fungal strains belonging to the class Sordariomycetes were isolated from soils collected from Gyeongsangbuk-do in Korea. They were identified as Cephalotrichum hinnuleum (UD CT 1-3-3 and KNU-19GWF1) and Thelonectria chlamydospora sp. nov. (UD ST 1-2-1). T. chlamydospora sp. nov. was morphologically identical to T. truncata, but its specific macroconidial dimensions, lower number of septations, and chlamydospore diameter render it distinct from the strains of the genus Thelonectria. The strains UD CT 1-3-3 and KNU-19GWF1 were developed flat, velvety to felty, and golden gray to brown-gray after 14 days of incubation at 25 °C on PDA. These strains were produced polyblastic conidiogenous cells and conidia were pale brown to brown, smooth, thin-walled, subglobose to ellipsoidal, arranged in chains, and the diameters of 6.7–9.0 × 3.7–5.1 μm. The strains were also confirmed by using the multi-locus genes using internal transcribed spacer (ITS) regions, partial large subunit (LSU), translation elongation factor 1α (TEF1-α), β-tubulin (TUB2), and actin (ACT) genes. This is the discovery of T. chlamydospora sp. nov. and Cephalotrichum hinnuleum, a new record from Korea.
1. Introduction
The phylum Ascomycota is comprised of extremely diverse fungal classes that play critical roles in the environment. It consists of 1,331 genera under 105 families, 32 orders, and six subclasses and class Sordariomycetes is the second largest [Citation1]. Morphologically, the heterogeneous group of fungi comprising saprobic and plant pathogenic species are classified under the family Microascaceae [Citation2]. The formation of dry-spored, indeterminate synnemata, and enteroblastic percurrent conidiogenesis are general characteristics of the genus Cephalotrichum, which belongs to the family Microascaceae, containing powdery conidia chains resembling a ‘bottle brush’ or ‘feather’, and comprised of their asexual states, but their teleomorph state is still unknown [Citation3]. However, for many years, the synonymity of Cephalotrichum and Doratomyces has been a subject of debate. Doratomyces was accepted as a separate genus, but Doratomyces is a synonym for Cephalotrichum [Citation4]. The genus Thelonectria is widespread and omnipresent fungi recently established hosts on woody within the family Nectriaceae; it was previously placed in the genus Neonectria [Citation5]. The genus Thelonectria is characterized by inconspicuous stromata; superficial, globose, subglobose, pyriform to elongated and smooth, or warty perithecia; a peritheal wall of 2 or 3 layers with prominent papilla; and smooth, hyaline, 1-septate ascospores. Members of this genus often cause small cancers and are found mostly on the bark of recently killed, dying, or diseased trees and rotting roots in tropical and subtropical regions [Citation6]. Recently, the presence of the cryptic species Thelonectria coronata and T. veuillotiana was examined, and it was discovered from temperate, tropical and subtropical regions. And the cryptic species were described using the methods of phylogenetic recognition through genealogical agreement between multiple phylogenies [Citation7]. Recently, T. veuillotiana was isolated from roots of a species of orchid (Oreorchis patens) from Korea [Citation8]. Ulleungdo is one of a small group of volcanic islands situated off the eastern coast of the Korean peninsula. Ulleungdo inhabits approximately 600 species of vascular plants, of which 39 are endemic to the island [Citation9]. The diversified fungal strains were recovered from agricultural soil in Ulleungdo such as Penicillium raphiae [Citation10], and Metarhizium guizhouense and Mortierella oligospora were also isolated from Dokdo nearest from Ulleungdo [Citation11].
The objective of the current study was to explore newly recorded fungal species from Gunwi and Ulleungdo based on cultural and morphological characteristics along with their molecular phylogeny. The three fungal strains were described and illustrated as a new record and one novel species from Korea.
2. Materials and methods
2.1. Sample collection
The soils were yellowish brown, contain plant debris and limited water efficiency nearby pine tree collected from Gunwi (36°11′45.9″N, 128°34′10.8″E) in 2018. The rhizospheric soils of root area of lantern (Campanula takesimana) and Ulleungdo stonecrop (Sedum takesimense) were collected from Ulleungdo (37°29′40.8″N, 130°49′39.4″E and 37°30′23.3″N, 130°49′5.9″E) of Gyeongsangbuk-do Province in 2018. Samples were stored in plastic bags at 4 °C until analysis. Soil serial dilutions were performed until a concentration of 10−3 was achieved, and 100 μL of each sample was spread on potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated for 2–3 days at 25 °C [Citation12]. Then, the pure cultures were transferred to new PDA plates and incubated at 25 °C for 5–7 days and cultured on a different media suitable for studying their cultural and morphological characteristics and conducting molecular analyses.
2.2. Morphological studies
Pure cultures were maintained to study the morphology of the three stains. The UD CT 1-3-3 and KNU-19GWF1 strains were grown on PDA and oatmeal agar (OA; Difco) for 14 days at 25 °C [Citation13]. According to Nirenberg [Citation14], synthetic nutrient agar (SNA: Agar-14.0 g/L, KH2PO4-1.0 g/L, KNO3-1.0 g/L, MgSO4.7H2O-0.25 g/L, KCl-0.5 g/L, Glucose-0.2 g/L, Sucrose-0.2 g/L) and PDA was used to study the strain UD ST 1-2-1 maintaining at 25 °C for 14 days [Citation15]. After incubation, the diameters of the colonies were measured, photographs were taken, and colony characteristics for each strain were recorded. A light microscope (BX-50; Olympus, Tokyo, Japan) was used to observe morphological characteristics.
2.3. Genomic DNA extraction, PCR, and sequencing
Genomic DNA was extracted from mycelia using the HiGene Genomic DNA prep kit (BIOFACT, Daejeon, Korea) according to the manufacturer’s instructions. For the strains UD CT 1-3-3 and KNU-19GWF1, the following target genes were amplified using internal transcribed spacer regions of rDNA including the 5.8S region (ITS1F or ITS5/ITS4) [Citation16,Citation17] and the partial large subunit of rDNA (LROR/LR5) [Citation18] as well as translation elongation factor 1α (TEF1-α) and β-tubulin (TUB2) genes using the primer pairs EF1-983F/EF1-2218R [Citation19] and BT2a/BT2b [Citation20], respectively. The ITS regions of rDNA including the 5.8S region and partial large subunit of rDNA (LSU) was also used to amplify the strain UD ST 1-2-1 along with TEF1-α (EF1-728F/EF1-986R), and actin (Tact1/Tact2) genes for molecular identification [Citation6]. The amplified PCR products were then purified with EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced (Macrogen, Daejeon, Korea).
2.4. Phylogenetic analysis
Strain sequences were compared with the additional sequences retrieved from the GenBank database of the National Center for Biotechnology Information (NCBI). The alignments of each gene were performed, and the sequences were combined to reveal the position of the strains on the phylogenetic tree using MEGA7.0 program. Evolutionary distance matrices were generated using Kimura’s neighbor-joining (NJ) algorithm model [Citation21]. To determine the exact taxonomic position of each strain, maximum likelihood (ML) and maximum parsimony (MP) trees were also constructed. In the neighbor-joining phylogenetic tree, nodes with filled circles represent maximum likelihood and maximum parsimony, and open circles indicate nodes corresponding to the maximum parsimony or maximum-likelihood algorithm. Phylogenetic analyses were performed using the MEGA 7.0 program with bootstrap values based on 1,000 replicates [Citation22].
3. Results
3.1. Taxonomy of UD CT 1-3-3
The strains UD CT 1-3-3 (NIBRFGC000505742) and KNU-19GWF1 (NREFFGC000000240) were studied and found same as well as clustered together with respect to molecular phylogeny. Thus, UD CT 1-3-3 and KNU-19GWF1 strains were identified as Cephalotrichum hinnuleum. Therefore, the cultural and morphological characteristics of the UD CT 1-3-3 strain were described only in this study.
Cephalotrichum hinnuleum Sand.-Den., Guarro & Gené, Studies in Mycology 83: 209 (2016) (, )
Figure 1. Cultural and morphological characteristics of UD CT 1-3-3. (A) Colonies on potato dextrose agar (PDA); (B) Colonies on oatmeal agar (OA) for 14 days of incubation at 25 °C; (C) Synnemata; (D) Apical portion of a synnema; (E) Polyblastic conidiogenous cells; (F) Conidia. Arrows indicate conidiogenous cells. Scale bars: C = 200 μm; D–F = 10 μm.
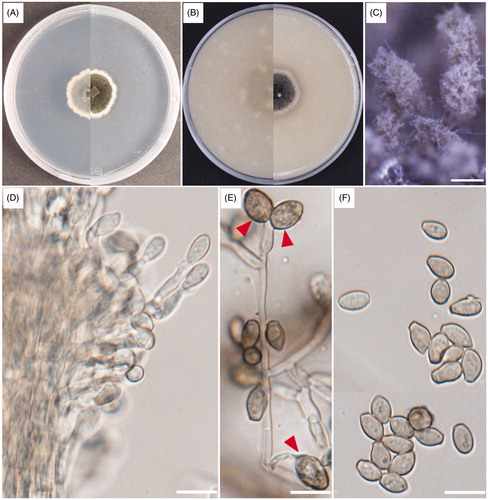
Table 1. Morphological characteristics of the NIBRFGC000505742 strain in reference to Cephalotrichum hinnuleum.
Cultural characteristics: The colonies of the strains UD CT 1-3-3 and KNU-19GWF1 were flat, velvety to felty, and golden gray to brown-gray on PDA after 14 days of incubation at 25 °C. The white margins of the colonies were regular and reached 22.0–27.0 mm. The reverse side showed a regular white margin and a brown-gray edge that became pale to dark gray in color (). The colonies grown on OA media were flat, velvety, and floccose with regular margins after 14 days of incubation at 25 °C. The reverse side was dark brown to olivaceous in color, and the colonies grew to 25.0–30.0 mm (). The cultural characteristics of both strains UD CT 1-3-3 and KNU-19GWF1 were studied on PDA and OA media and found the same characteristics.
Morphological characteristics: Morphological characteristics were observed using the strain UD CT 1-3-3 grown on PDA for 14 days at 25 °C. The strain UD CT 1-3-3 produced numerous dark-brown to black-colored synnemata, compact stipes, and light gray to gray conidial heads. They were clavate to ellipsoidal, setae-absent, 700–1,400 μm in height, and 10–28 μm wide (). The hyphae were septate, hyaline, light brown, smooth, thin-walled, and 2–3 μm wide. The conidiophores were branched, septate, hyaline, pale brown, smooth, and thin-walled and usually aggregated with the dense synnemata (). This strain also produced polyblastic conidiogenous cells (). The conidia were pale brown to brown, smooth, thin-walled, and subglobose to ellipsoidal with shortened bases and blunted apexes and arranged in chains with diameters of 6.7–9.0 × 3.7–5.1 μm () ().
3.2. Phylogenetic analysis of UD CT 1-3-3
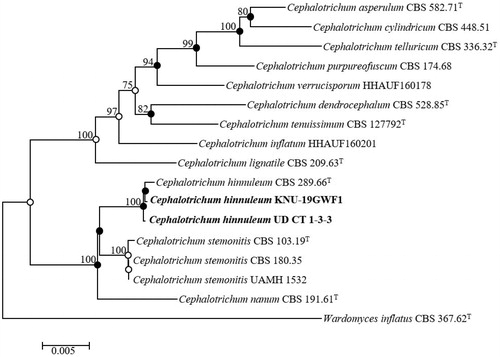
Four multi-genes were amplified to identify the UD CT 1-3-3 and KNU-19GWF1 strains. The sequences from ITS regions (569 bp, 585 bp), LSU (858 bp, 820 bp), TEF1-α (940 bp, 893 bp), and TUB2 (541 bp, 518 bp) were obtained from both strains. The obtained sequences were submitted in NCBI GenBank and accession numbers were for ITS (LC509451, LC519564), LSU (LC509453, LC519565), TEF1-α (LC509454, LC519562), TUB2 (LC519561, LC519563) from UD CT 1-3-3 and KNU-19GWF1 strains, accordingly (). BLAST search results revealed that the UD CT 1-3-3 strain showed the highest similarities (100%) with the previously identified C. hinnuleum CBS 289.66T (previously known as Doratomyces stemonitis) based on ITS regions. LSU and TEF1-α showed maximal similarities (99.64% and 99.78%) with the C. hinnuleum CBS 289.66T, respectively. And TUB2 also showed maximum similarities (99.82%) with the C. hinnuleum CBS 289.66T.
Table 2. List of species used in this study and their GenBank accession numbers for phylogenetic analysis.
The KNU-19GWF1 strain also showed maximum similarities with ITS regions (100%), LSU (100%), TEF1-α (99.78%), and TUB2 (99.81%) with the previously identified C. hinnuleum CBS 289.66T strain. The deposited sequences of existing Cephalotrichum species in GenBank were used to construct the taxonomic position of KNU-19GWF1 in the phylogenetic tree (). The neighbor-joining phylogenetic tree revealed through the alignment of their ITS regions including 5.8S of the nuclear ribosomal DNA region, LSU, TUB2, and TEF1-α gene sequences that the position of UD CT 1-3-3 was closely clustered with the C. hinnuleum CBS 289.66T () showing strong bootstrap values of 100%. These results indicate that UD CT 1-3-3 and KNU-19GWF1 strains are, in fact, Cephalotrichum hinnuleum. Neighbor-joining, maximum-likelihood, and maximum parsimony trees were constructed to determine the precise taxonomic location of the strains and are indicated with nodes in neighbor-joining phylogenetic tree. Filled circles indicate that the corresponding nodes were recovered in trees generated with the maximum-likelihood and maximum parsimony algorithms. Open circles indicate that the corresponding nodes were also recovered from maximum-likelihood or maximum parsimony algorithms. The exact taxonomic position of the strain was determined through the analysis of the maximum parsimony (tree length = 434, consistency index = 0.64, retention index = 0.82, and composite index = 0.63) ().
Figure 2. Neighbor-joining phylogenetic tree of UD CT 1-3-3 and KNU-19GWF1 based on the combined sequences (ITS + LSU+TUB2+TEF1-α), showing the relationships between Cephalotrichum hinnuleum and the closest Cephalotrichum spp. Wardomyces inflatus CBS 367.62T was used as an outgroup. The numbers above the branches represent the bootstrap values (>70%) obtained for 1,000 replicates. The isolated strains of this study are indicated in bold. Bar, 0.005 substitutions per nucleotide position.
3.3. Taxonomy of UD ST 1-2-1
The strain UD ST 1-2-1 showed different morphological characteristics when compared to other allied species. Therefore, it was defined as a new species. The description includes the cultural and asexual morph features, the sexual morph features were undetermined from the cultural media (PDA and SNA).
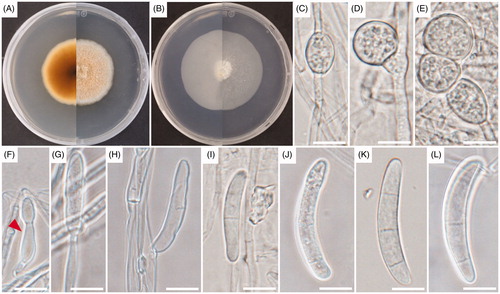
Thelonectria chlamydospora K. Das, S.Y. Lee and H.Y. Jung, sp. nov. ()
Figure 3. Cultural and morphological characteristics of UD ST 1-2-1. (A) Colonies on potato dextrose agar (PDA); (B) Colonies on synthetic nutrient agar (SNA) for 14 days of incubation at 25 °C; (C–E) Chlamydospores; (F) Phialidic cell; (G) Conidiophore; (H) Conidiogenous cell; (I–L) Macroconidia. Arrows indicate phialidic cells. Scale bars: C–L = 10 μm.
MycoBank: MB834497
Etymology: The specific epithet refers to the Greek term chlamydos-, cloak, and the Latin term spora, spore.
Typus: Ulleungdo, Gyeongsangbuk-do (37°30′23.3″N, 130°49′5.9″E), Korea, isolated from Ulleungdo stonecrop (Sedum takesimense) rhizospheric soil. The stock culture (NIBRFGC000500679 = KCTC 56672) was deposited in the National Institute of Biological Resources (NIBR) and Korean Collection for Type Cultures (KCTC), metabolically inactive culture.
Habitat and distribution: Rhizospheric soil regions of Ulleungdo stonecrop (Sedum takesimense) in South Korea.
Cultural characteristics: Colonies on PDA formed floccose aerial mycelium that were white to saffron; the colony was saffron to white color in reverse side and reached 42.0–43.0 mm after 14 days of incubation at 25 °C with even margin expansion (). Colonies on SNA media were slightly floccose and sparse white to light brown. The colony reverse side was white and grew to 60.0–66.0 mm in diameter after 14 days of incubation at 25 °C ().
Morphological characteristics: Sexual morph: undetermined. Asexual morph: micromorphological characteristics were studied by culturing on SNA media maintaining at 25 °C for 14 days. Mycelia were septate branched, hyphae hyaline, light brown, and smooth with a diameter between 1.9–3.9 μm. Chlamydospores observed in culture were hyaline and globose to sub-globose with the diameters between 10.8–21.8 × 5.6–16.7 μm (). Phialides were borne apically with clusters of cells in branch irregularly, alone, or developed from hyphae directly; they were curved, cylindrical, or slightly swollen with diameters between 12.8–14.8 × 3.0–4.3 μm (). The produced conidiophores were branched or unbranched, hyaline, light brown, and gave rise to conidiogenous cells (). The conidiogenous cells were septate with diameters of 10.8–30.7 × 2.4–4.9 μm (average = 18.1 × 3.7 μm) (). The UD ST 1-2-1 strain generated a lower quantity of macroconidia on artificial media. On SNA, Macroconidia were hyaline, produced slimy droplets in aerial mycelium or on the agar surface. Macroconidia were cylindrical to slightly fusiform, thick-walled, curved, and rounded on both ends; 1–3‐septate formed diameters of 28.6–40.4 × 4.8–6.8 μm (average = 37.6 × 5.9 μm), one septate (28.6–40.3 × 4.8–5.7 μm), and three septate (37.2–40.4 × 5.5–6.8 μm) (). Microconidia were not observed in culture.
Note: The UD ST 1-2-1 strain produced conidiogenous cells with nearly similar diameters (10.8–30.7 × 2.4–4.9 μm) with the nearest known species of T. truncata (14.0–30.0 × 2.5–4.5 μm) [Citation15]; whereas, conidiogenous cells diameter were not mentioned for another strain of T. truncata () [Citation7]. The UD ST 1-2-1 strain produced smaller, curved, cylindrical, or slightly swollen phialides (12.8–14.8 × 3.0–4.3 μm), and T. truncata produced phialides that were cylindrical or sometimes slightly swollen 16.0–20.6 × 3.5–4.4 μm [Citation7]. The UD ST 1-2-1 strain generated fewer, cylindrical to slightly fusiform, thick wall, and curved macroconidia that were rounded on both ends:1–3–septate: 28.6–40.4 × 4.8–6.8 μm, one septate 28.6–40.3 × 4.8–5.7, and three septate 37.2–40.4 × 5.5–6.8 μm (mean 37.6 × 5.9 μm). In contrast, the closest species T. truncata produced macroconidia that were cylindrical to slightly fusiform with curved or rounded tips, 3–5(–6)–septate: three–septate 46.9–58.9 × 4.9–5.8 μm (mean 53.3 × 5.4 μm), four–septate 55–67.5 × 5.0–6.0 μm (mean 61.2 × 5.5 μm), and five–septate 65.4–77.0 × 5.2–6.3 μm (mean 71.2 × 5.8 μm) [Citation7]. Moreover, another strain of T. truncata produced macroconidia that were cylindrical to slightly fusiform and curved with the rounded ends, 3–5(–6)–septate, 42.0–62.0 μm [Citation15]. The UD ST 1-2-1 strain also generated numerous chlamydospores that were observed when cultured on SNA media; they were hyaline, globose to subglobose, and formed diameters of 10.8–21.8 × 5.6–16.7 μm. On the other hand, the closest known type species of T. truncata does not produce chlamydospores on SNA media [Citation7], similar to another strain of T. truncata [Citation15]. Also, there was a difference in the number of septate macroconidia with the closest species. The closest species produced 3–5(–6) septate macroconidia, but the UD ST 1-2-1 strain developed 1–3–septate macroconidia. A comparison between the nearest certain species of the genus showed a lower number of septations in macroconidia, smaller macroconidial size, and chlamydospores. Therefore, the morphology of the UD ST 1-2-1 strain is totally different from previously identified species of Thelonectria.
Table 3. Morphological characteristics of Thelonectria chlamydospora sp. nov. and a comparison with the closest species from the genus Thelonectria.
3.4. Phylogenetic analysis of UD ST 1-2-1
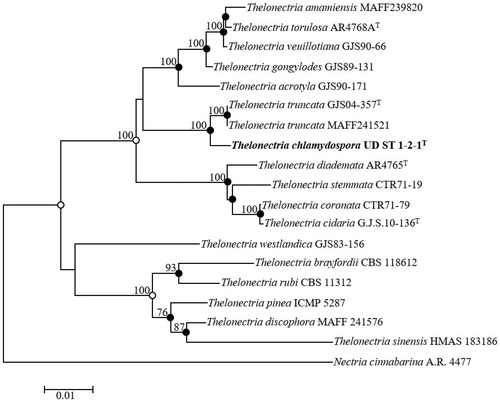
Sequences for 563 bp of the ITS region, 843 bp of LSU, 307 bp of TEF1-α, and 694 bp of ACT were obtained from the UD ST 1-2-1 strain. The obtained sequences from UD CT 1-3-3 was deposited in NCBI GenBank and accession numbers were LC509450, LC509452, LC519559, LC519560 from ITS, LSU, TEF1-α, and ACT, accordingly. ITS regions showed maximum similarities with the Thelonectria truncata CBS 132329T (98.43%) and T. truncata 9386 (98.50%). LSU and TEF1-α showed maximum similarities with the strains T. truncata CBS 132329T (99.15%) and T. veuillotiana G.J.S.09-407 (86.75%), respectively. And the ACT gene also showed maximum similarities with the T. truncata CBS 132329T (99.16%). Existing Thelonectria species sequences retrieved from GenBank were used to construct the phylogenetic tree (). The phylogenetic tree showed that the position of UD ST 1-2-1 was distinctly clustered in a different clade with respect to the previously identified strains of T. truncata (). Thus, the UD ST 1-2-1 strain is phylogenetically distinct from the other Thelonectria species. The combination of ITS regions, LSU, TEF1-α, and ACT gene sequences clarified the boundaries between the 18 strains of Thelonectria. The exact taxonomic position of the UD ST 1-2-1 strain was indicated by the node in the neighbor-joining phylogenetic tree along with the filled nodes in the maximum likelihood and maximum parsimony trees. The corresponding nodes were also recovered using the maximum likelihood or maximum parsimony algorithms, as indicated by the open circles. The combination of the sequences was used for the phylogenetic analysis based on maximum parsimony (tree length = 578, consistency index = 0.63, retention index = 0.80, and composite index = 0.58) to determine the taxonomic position of the strain UD ST 1-2-1 ().
Figure 4. Neighbor-joining phylogenetic tree of UD ST 1-2-1 based on the combined sequences (ACT+LSU + ITS+TEF1-α), showing the relationships between Thelonectria chlamydospora sp. nov. and the closest Thelonectria spp. Nectria cinnabarina A.R. 4477 was used as an outgroup. The numbers above the branches represent the bootstrap values (>70%) obtained for 1,000 replicates. The isolated strain of this study is indicated in bold. Bar, 0.01 substitutions per nucleotide position.
4. Discussion
In this study, the three strains were collected from rhizospheric soils regions in Gunwi and Ulleungdo, Korea: KNU-19GWF1, UD CT 1-3-3, and UD ST 1-2-1. The strains were identified as Cephalotrichum hinnuleum (UD CT 1-3-3 and KNU-19GWF1) and Thelonectria chlamydospora sp. nov. (UD ST 1-2-1).
From the previous studies, the most of the members of Cephalotrichum found on decaying plant materials, straw, dung, wood, and soil [Citation4]. However, C. microsporum has been found on indoor substrates like wood or in man-made environments [Citation23], and C. purpureofuscum has been found in indoor air [Citation3,Citation13]. C. stemonitis was identified in coyote and rat dung, the indoor air of a honeybee (Apis mellifera) overwintering facility, the cones of white spruce, sandy soil, decayed wood of white spruce, soil of elm woods, and agricultural soil [Citation24]. Recently, there are 14 new species of Cephalotrichum were explained based on the isolation from soils in China [Citation25]. Although Cephalotrichum species are not considered as human pathogens and also not known to produce mycotoxins, but C. gorgonifer was collected from human clinical samples and can survive at human body temperatures [Citation13]. The type species C. hinnuleum was isolated from dung of deer in Australia [Citation13], whereas, the strains identified as C. hinnuleum (UD CT 1-3-3 and KNU-19GWF1) isolated from rhizospheric soils of root area of lantern (Campanula takesimana) and pine tree from Ulleungdo in Korea. Previously, the fungal strains Penicillium sanguifluum and P. pasqualense were collected under the rhizosphere of a pine tree located Dokdo Island, Korea [Citation26], whereas, P. piscarium and Talaromyces versatilis reported from freshwater environment [Citation27].
The earliest record of Thelonectria (T. discophora), previously considered as cosmopolitan, arises from Chile [Citation28]. The T. discophora complex occured with the diversified habitats and plant substrates such as twig of bark and recently dead or dying trees branches [Citation29]. Currently, 9 species within the genus Thelonectria have been identified and five of which were previously reported from China [Citation6]. T. coronata (as Nectria coronata) was recently isolated from Hong Kong [Citation30], T. jungneri (Henn.) and T. lucida (as Neonectria lucida) were later reported from the Taiwan Province [Citation29], and T. veuillotiana (as Neonectria veuillotiana) was found in the Hubei Province [Citation31]. Also, T. coronata and T. veuillotiana were sufficient amount in temperate, tropical, and subtropical regions and grow on the bark of dead or dying trees recently and on canker lesions caused with the other causal agents [Citation29,Citation31]. Though, the different fungal strains under the genus Thelonectria were isolated from diversified habitats, plant substrates along with plants parts from temperate, tropical, and subtropical regions in different countries, but our strain UD ST 1-2-1 was identified from rhizospheric soil regions of Ulleungdo stonecrop (Sedum takesimense) from Ulleungdo in South Korea. In future study, the taxonomy requires further study with the wide ranges of geographical location along with their pathogenicity tests based on the hosts.
Further investigations are necessary to explore the etiology of C. hinnuleum and T. chlamydospora based on the host(s), environments and agricultural land conditions in Korea. Based on their cultural and morphological characteristics and phylogenetic analyses, the UD CT 1-3-3 and KNU-19GWF1 strains were similar to C. hinnuleum. The UD ST 1-2-1 strain was different from other identified species under the genus Thelonectria, and it was described as a novel species. Thus, the newly discovered species was proposed as Thelonectria chlamydospora sp. nov. and Cephalotrichum hinnuleum, a new record from Korea.
Disclosure statement
The authors declare that they have no potential conflicts of interest.
Additional information
Funding
References
- Maharachchikumbura SSN, Hyde KD, Jones EB, et al. Families of Sordariomycetes. Fungal Divers. 2016;79(1):1–317.
- Malloch D. New concepts in the Microascaceae illustrated by two new species. Mycologia. 1970;62(4):727–740.
- Abbott SP. Holomorph studies of the Microascaceae [Ph.D. dissertation]. Canada: University of Alberta; 2000.
- Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. 2nd ed. Eching. Germany: IHW Verlag; 2007.
- Rossman AY, Samuels GJ, Rogerson CT, et al. Genera of bionectriaceae, hypocreaceae and nectriaceae (Hypocreales, Ascomycetes). Stud Mycol. 1999;42:1–248.
- Chaverri P, Salgado-Salazar C, Hirooka Y, et al. Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol. 2011;68:57–78.
- Salgado-Salazar C, Rossman A, Samuels GJ, et al. Multigene phylogenetic analyses of the Thelonectria coronata and T. veuillotiana species complexes. Mycologia. 2012;104(6):1325–1350.
- Lee BH, Kim DY, Park H, et al. Notes on endophytic fungi isolated from roots of Oreorchis patens in Korea. Kor J Mycol. 2016;44:184–187.
- Sun BY, Shin H, Hyun JO, et al. Vascular plants of Dokdo and Ulleungdo islands in Korea. Incheon, Korea: National Institute of Biological Resources; 2014.
- Paul NC, Mun HY, Lee HW, et al. A new record of Penicillium raphiae isolated from agricultural soil of Ulleung Island, Korea. Mycobiology. 2014;42(3):282–285.
- Lee HY, Nguyen TTT, Mun HY, et al. Confirmation of two undescribed fungal species from Dokdo of Korea based on current classification system using multi loci. Mycobiology. 2015;43(4):392–401.
- Lee SH, Park HS, Nguyen TTT, et al. Characterization of three species of Sordariomycetes isolated from freshwater and soil samples in Korea. Mycobiology. 2019;47(1):20–30.
- Sandoval-Denis M, Guarro J, Cano-Lira1 JF, et al. Phylogeny and taxonomic revision of Microascaceae with emphasis on synnematous fungi. Stud Mycol. 2016;83:193–233.
- Nirenberg HI, Aoki T. Fusarium nisikadoi, a new species from Japan. Mycoscience. 1997;38(3):329–333.
- Zeng ZQ, Zhuang WY. Three new Chinese records of Nectriaceae. Mycosystema. 2016;35:1399–1405.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97(1):84–98.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Flannigan B, Samson RA, Miller JD (eds.). Microorganisms in home and indoor work environments. Diversity, health impacts, investigation and control. 2nd ed. Florida, USA: CRC press; 2011.
- Sabev HA, Handley PS, Robson GD. Fungal colonization of soil-buried plasticized polyvinyl chloride (pPVC) and the impact of incorporated biocides. Microbiology. 2006;152(Pt 6):1731–1739.
- Jiang YL, Xu JJ, Wu YM, et al. Studies on Cephalotrichum from soils in China–twelve new species and two new combinations. Mycotaxon. 2011;117(1):207–225.
- Pangging M, Nguyen TTT, Lee HB. New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology. 2019;47(2):154–164.
- Heo I, Hong K, Yang H, et al. Diversity of Aspergillus, Penicillium, and Talaromyces species isolated from freshwater environments in Korea. Mycobiology. 2019;47(1):12–19.
- Brayford D, Honda BM, Mantiri FR, et al. Neonectria and Cylindrocarpon: the Nectria mammoidea group and species lacking microconidia. Mycologia. 2004;96(3):572–597.
- Hsieh HM, Chou JC, Ju YM. Nectriaceous fungi collected from forests in Taiwan. Bot Stud. 2020;61(1):187–203.
- Lu BS, Hyde KD, Ho WH. Checklist of Hong Kong fungi. Fungal Diversity Series 5. Hong Kong: Fungal Diversity Press; 2000.
- Zhuang WY, Nong Y, Luo J. New species and new Chinese records of bionectriaceae and nectriaceae (Hypocreales, Ascomycetes) from Hubei, China. Fungal Divers. 2007;24:347–357.
- Carlucci A, Lops F, Mostert L, et al. Occurrence fungi causing black foot on young grapevines and nursery rootstock plants in Italy. Phytopathol Mediterr. 2017;56:10–39.
- Zeng ZQ, Zhuang WY. The genera Rugonectria and Thelonectria (Hypocreales, Nectriaceae) in China. MycoKeys. 2019;55:101–120.
- Zeng ZQ, Zhuang WY. Four new taxa of Ilyonectria and Thelonectria (Nectriaceae) revealed by morphology and combined ITS and β-tubulin sequence data. Phytotaxa. 2013;85(1):15–25.
- Salgado-Salazar C, Rossman AY, Samuels GJ, et al. Phylogeny and taxonomic revision of Thelonectria discophora (Ascomycota, Hypocreales, Nectriaceae) species complex. Fungal Divers. 2015;70(1):1–29.
