Abstract
Scopulariopsis brevicaulis is a widely distributed soil fungus known as a common saprotroph of biodegradation. It is also an opportunistic human pathogen that can produce various secondary metabolites. Here, we report the first complete mitochondrial genome sequence of S. brevicaulis isolated from air in South Korea. Total length of the mitochondrial genome is 28,829 bp and encoded 42 genes (15 protein-coding genes, 2 rRNAs, and 25 tRNAs). Nucleotide sequence of coding region takes over 26.2%, and overall GC content is 27.6%. Phylogenetic trees present that S. brevicaulis is clustered with Lomentospora prolificans with presenting various mitochondrial genome length.
Scopulariopsis genus contains 22 fungal species [Citation1] usually isolated from soil, air, plant debris, paper, and moist indoor environments [Citation2,Citation3]. They are soil saprophytes, pathogenic to animals including human [Citation4]. Scopulariopsis brevicaulis (teleomorph: Microascus brevicaulis), which is one of five human pathogenic Scopulariopsis species usually involved in onychomycosis [Citation5–7], can cause various skin problems and onychomycosis [Citation7]. Here, we present a complete mitochondrial genome of S. brevicaulis as the first mitochondrial genome in Scopulariopsis genus.
A strain of S. brevicaulis was collected from mycobiota of air inside and outside the Meju fermentation room in South Korea [Citation8] and its total DNA was extracted by using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). In total 2.825 Gbp raw data generated by MiSeq () were subject to de novo assembly done by Velvet 1.2.10 [Citation9] and gap filling with SOAPGapCloser 1.12 [Citation10] to get complete mitochondrial genome after confirming each bases using BWA 0.7.17 and SAMtools 1.9 [Citation11,Citation12]. Coverage of raw reads against the mitochondrial genome is 3090.52× (). Geneious R11 v11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its mitochondrial genome by comparing with that of Collectotrichum siamense strain YT02 (KX885103). ARWEN [Citation13] was used for finding novel tRNAs on the mitochondrial genome. The sample was deposited into Korean Agricultural Culture Collection (KACC-47468).
Table 1. Raw reads generated by MiSeq for assembling mitochondrial genome of Scopulariopsis brevicaulis KACC-47468.
The length of S. brevicaulis mitochondrial genome (GenBank accession number: MK672942) is 28,829 bp (). S. brevicaulis mitochondrial genome encodes 42 genes consisting of 15 protein-coding genes, 2 rRNAs, and 25 tRNAs (). This configuration of S. brevicaulis mitochondrial genome is same to those of C. siamense used for mitochondrial genome annotation and Lomentospora prolificans, which is the nearest species in phylogenetic tree (). Nucleotide sequence of coding region takes over 26.2%, and overall GC content is 27.6%. It is different from those of L. prolificans: GC ratio of coding region is 25.5% and overall GC content is 26.9%.
Figure 1. Complete map of mitochondrial genome of Scopulariopsis brevicaukis KACC-47468. Gray graph inside circular diagram presents GC ratio of mitochondrial genome. Colorful bars on outer circular form indicate PCGs (yellow, pink, green, and purple), tRNAs (dark blue), and rRNAs (red).
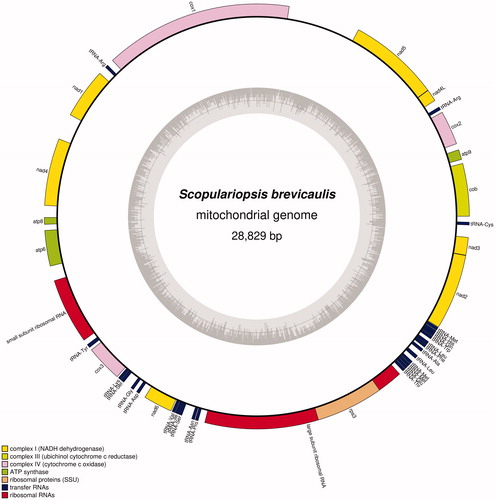
Figure 2. (A) Maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (boostrap repeat is 10,000) phylogenetic trees of twenty Microascales mitochondrial genomes including that of Scopulariopsis brevicaulis and Aspergillus mitochondrial genomes as an outgroup: Scopulariopsis brevicaulis (MK672942 in this study) Lomentospora prolificans (NLAX01001625), Ophiocordyceps sinensis (KY622006), Fusarium mangiferae (KP742838), Annulohypoxylon stygium (MH620792), Juglanconis juglandina (KY575057), Chrysoporthe austroafricana (KT380883), Verticillium dahliae (DQ351941), Colletotrichum lupini (KT918406), Colletotrichum tamarilloi (KU196965), Colletotrichum acutatum (KR349346), Colletotrichum fioriniae (KU375885), Colletotrichum salicis (KY774449), Colletotrichum lindemuthianum (MF595869), Colletotrichum gloeosporioides (KX885104), Colletotrichum siamense (KX885103), Colletotrichum aenigma (KX885105), and Colletotrichum fructicola (KX034082), Endoconidiophora resinifera (MK026449), and Aspergillus flavus (JQ355000). Phylogenetic tree was drawn based on maximum likelihood phylogenetic tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining methods, respectively. (B) Gray graph displays length of twenty mitochondrial genomes.
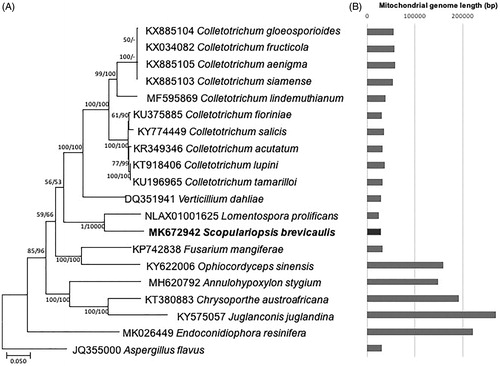
Each sequence alignment of conserved 12 genes from 20 Microascales mitochondrial genomes by MAFFT 7.450 [Citation14] was concatenated. Annotation of L. prolificans mitochondrial genome was also conducted for incorporating into the trees. The bootstrapped neighbor-joining and maximum likelihood phylogenic trees were constructed using MEGA X [Citation15]. Lengths of nineteen Microascales mitochondrial genomes vary, ranging from 23,987 bp (L. prolificans) to 2,67,504 bp (Juglanconis juglandina; ). Phylogenetic tree presents that S. brevicaulis is clustered with L. prolificans, of which lengths are two of the shortest mitochondrial genomes (). Interestingly, length of Ophiocordyceps sinensis mitochondrial genome is much larger than that of Fusarium mangifere but both were clustered in one clade with high bootstrap values (). The mitochondrial genome expansion in Aspergillus genus [Citation16–19] and variable size of Fusarium mitochondrial genomes [Citation18,Citation20,Citation21] suggested that mitochondrial genome expansion events may be occurred several times during evolution of fungal species. In addition, tree displayed two distinct clades in Colletotrichum genus, which is congruent with previous phylogenetic study [Citation22] (). This mitochondrial genome as a first reported Scopulariopsis mitochondrial genome will be a useful molecular resource to understand mitochondrial genome characteristics. Also, since the fungus is an opportunistic human pathogen, the mitochondrial genome information can be used to study of the host-pathogen interaction because mitochondrial activity could be involved in its pathogenic properties.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MK672942 in NCBI GenBank.
Additional information
Funding
References
- Kirk P, Cannon P, Minter D, et al. Dictionary of the Fungi. 10th ed. Wallingford, UK; 2008.
- Kwon-Chung K, Bennett J. Infections due to miscellaneous molds. In: Kwon-Chung K, Bennett J, editors. Medical mycology. Philadelphia, PA: Lea & Febiger; 1992. p. 733–739.
- Samson R, Houbraken J, Thrane U, et al. Food and indoor fungi. CBS laboratory manual series 2. Utrecht: CBS-Fungal Biodiversity Centre; 2010.
- Issakainen J, Heikkilä H, Vainio E, et al. Occurrence of Scopulariopsis and Scedosporium in nails and keratinous skin. A 5-year retrospective multi-center study. Med Mycol. 2007;45(3):201–209.
- Summerbell R, Kane J, Krajden S. Onychomycosis, tinea pedis and tinea manuum caused by non-dermatophytic filamentous fungi. Mycoses. 1989;32(12):609–619.
- Tosti A, Piraccini B, Stinchi C, et al. Onychomycosis due to Scopulariopsis brevicaulis: clinical features and response to systemic antifungals. Br J Dermatol. 1996;135(5):799–802.
- Cuenca-Estrella M, Gomez-Lopez A, Mellado E, et al. Scopulariopsis brevicaulis, a fungal pathogen resistant to broad-spectrum antifungal agents. Antimicrob Agents Chemother. 2003;47(7):2339–2341.
- Kim D-H, Kim S-H, Kwon S-w, et al. The mycobiota of air inside and outside the meju fermentation room and the origin of meju fungi. Mycobiology. 2015;43(3):258–265.
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, et al. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 2011;12(Suppl 14):S2.
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079.
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2. 2013.
- Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24(2):172–175.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549.
- Joardar V, Abrams NF, Hostetler J, et al. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 2012;13(1):698.
- Xu Z, Wu L, Liu S, et al. Structure characteristics of Aspergillus egyptiacus mitochondrial genome, an important fungus during the fermentation of dark tea. Mitochondrial DNA Part B. 2018;3(2):1135–1136.
- Park J, Kwon W, Huang X, et al. Complete mitochondrial genome sequence of a xerophilic fungus Aspergillus pseudoglaucus. Mitochondrial DNA Part B. 2019;4(2):2422–2423.
- Park J, Kwon W, Kim J-B, et al. Complete mitochondrial genome sequence of lettuce pathogenic fungus, Fusarium oxysporum f. sp. lactucae 09-002. Mitochondrial DNA Part B. 2019;4(2):3434–3436.
- Brankovics B, van Dam P, Rep M, et al. Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics. 2017;18(1):735.
- Kwon W, Park J, Kim J-B, et al. Complete mitochondrial genome sequence of lettuce pathogenic fungus, Fusarium oxysporum f. sp. lactucae 16-086. Mitochondrial DNA Part B. 2019;4(2):3227–3228.
- Chen Y, Qiao W, Zeng L, et al. Characterization, pathogenicity, and phylogenetic analyses of colletotrichum species associated with brown blight disease on Camellia sinensis in China. Plant Dis. 2017;101(6):1022–1028.
