Abstract
A cordycipitoid fungus infecting Hepialidae sp. in Nepal was supposed to be identical to Cordyceps liangshanensis, originally described from southwestern China, and thus, transferred to the genus Metacordyceps or Papiliomyces in previous studies. However, our multi-gene (nrSSU-nrLSU-tef-1α-rpb1-rpb2) phylogenetic and morphological studies based on the type specimen and additional collections of C. liangshanensis revealed that the fungus belongs to the genus Ophiocordyceps (Ophiocordycipitaceae). Therefore, a new combination O. liangshanensis was made, and a detailed description of this species was provided.
1. Introduction
Cordyceps liangshanensis M. Zang, D.Q. Liu & R.Y. Hu is well-known in southwestern China and has been used as a Traditional Chinese Medicine (TCM) named “Mai-Gan-Chong-Cao” for a long time. Like Ophiocordyceps sinensis (Berk.) G.H. Sung et al., C. liangshanensis parasitizes soil-borne larvae of Hepialidae sp., and has a limited distribution in China [Citation1]. The caterpillar-fungus resulting from fungal parasitism has been frequently used for the treatment of chronic cough, hemoptysis, asthma, lumbago, impotence and seminal emissions, and other diseases in Yi Nationality areas as it has special effects on cough expectorant, reinforcing kidney, nourishing lung, etc. [Citation1,Citation2]. The main ingredients of the natural C. liangshanensis are similar to the those of the natural O. sinensis, which contains amino acids, mannitol, adenosine, ergosterol, stearic acid, alkaloids, and organic acids [Citation1,Citation3]. The contents of polysaccharides, flavonoids, and nucleosides of C. liangshanensis were a little bit lower than those of O. sinensis, while the mannitol and saponins were higher than those of O. sinensis [Citation3]. It has been long recognized as a prized medicinal fungus and a desirable alternative for natural O. sinensis by local people. This medicinal fungus was firstly recorded in “Sichuan Tongzhi,” and it was treated as a new species and named Cordyceps liangshanensis by Zang et al. [Citation4]. The medicinal virtue of C. liangshanensis is also highly valued by herbalists. This fungus was included in the standardized herbal medicines of Sichuan (enlarged edition) in 1987.
With the rapid development of molecular phylogenetic techniques, considerable changes to the taxonomy of Cordyceps s. l. have occurred. Sung et al. [Citation5] proposed the genus Metacordyceps to accommodate some species of Cordyceps s. l., which was characterized by solitary or grouped stromata which are simple or branched, with a fleshy or tough whitish stipe, a greenish yellow to greenish cylindrical to enlarged fertile part, and perithecia partially or completely immersed in stromata.
Specimens collected from Nepal were once regarded as Cordyceps liangshanensis and, subsequently this species was moved from Cordyceps to the genus Metacordyceps based on multi-gene phylogenetic evidence [Citation5]. Based on the DNA sequences generated by Sung et al. [Citation6], C. liangshanensis was transferred to Papiliomyces in the family Clavicipitaceae. However, the type or authentic materials of C. liangshanensis were not examined in the aforementioned studies. There is a need to reinvestigate the type and additional collections of C. liangshanensis.
In this study, the type and other specimens from type locality of C. liangshanensis were collected and examined. The redescription was carried out on the basis of five-gene (nrSSU, nrLSU, tef-1α, rpb1, and rpb2) molecular phylogenetic analysis and morphological observations. Our data indicated that collections of C. liangshanensis differ from those from Nepal generated by Sung et al. [Citation5], and the species transferred to Metacordyceps or Papiliomyces was not justified.
2. Materials and methods
2.1. Specimens
The type specimen of Cordyceps liangshanensis (KUN-HKAS 7723) was borrowed from Kunming Institute of Botany, Chinese Academy of Sciences. Additional collections were made in two locations in southwestern China, one location in Leibo County, Sichuan Province, and another in Shuifu County, Yunnan Province. Specimens were stored in plastic containers at low temperature and transported to the laboratory for identification and isolation. Afterward, they were deposited at Yunnan Herbal Herbarium (YHH), Yunnan University.
2.2. Fungal isolation and culture
Specimens were rinsed with tap water, and then washed with sterile distilled water. For the purpose of obtaining pure cultures, stromata were immersed in 30% H2O2 for 5 min, rinsed with sterile water, and then dried on sterilized filter paper. Stromata were then cut off and a small piece of tissue was inoculated onto potato dextrose agar (PDA: fresh diced potato 200 g, dextrose 20 g, agar 18 g, in 1000 ml distilled water) plates. The purified fungal strains were maintained in a culture room at 25 °C or transferred to PDA slants and stored at 4 °C, and were deposited to the Yunnan Fungal Culture Collection (YFCC) at the Institute of Herb Biotic Resources of Yunnan University.
2.3. Morphological observations
Specimens were examined in the laboratory using the Canon 750 D camera (Canon Inc., Tokyo, Japan) and Olympus SZ60 stereo dissecting microscope (Olympus Corporation, Tokyo, Japan). Cultures on PDA slants were transferred to PDA plates and incubated at 25 °C for 2 months. The colors of fresh specimens and cultures were characterized by the color standard [Citation7]. Frozen sections and glass slides with lactic acid phenol cotton blue solution were prepared for morphological observation and measurement of sexual morph under a light microscope (BX53, Olympus Corporation, Tokyo, Japan). Morphological description of asexual morph was conducted as the method described by Wang et al. [Citation8]. Micro-morphological observations and measurements were performed using the Olympus BX53 stereomicroscope and a scanning electron microscope (Quanta 200 FEG, FEI Company, Hillsboro, USA).
2.4. DNA extraction, PCR, and sequencing
Specimens and live axenic cultures were prepared for DNA extraction. Genomic DNA was extracted using a Genomic DNA Purification Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s protocol. The primers used for PCR amplification of nrSSU, nrLSU, tef-1α, rpb1, and rpb2 are listed in . All PCR reactions were performed in a final volume of 50 μl containing 25 μl 2 × Taq PCR Master Mix (Tiangen Biotech Co., Ltd, Beijing, China), 0.5 μl of each primer (10 μM), 1 μl of genomic DNA, and 23 μl of RNase-free water. Target gene amplification and sequencing were performed according to the methods described in our previous publication [Citation9].
Table 1. PCR primers used in this study.
2.5. Phylogenetic analysis
Five gene sequences were retrieved from GenBank, and combined with those generated in our study (). The sequences were aligned using the programs Clustal X 2.0 and MEGA v6.06 [Citation13,Citation14]. After sequence alignment, the aligned sequences of five genes were concatenated. Partition homogeneity test was conducted using PAUP* 4.0b10 [Citation15], and the result revealed that there was no significant conflict among different data partitions. Program PartitionFinder V1.1.1 identified eleven data partitions, one each for nrSSU and nrLSU, and nine for each of the three codon positions for the protein coding genes tef-1α, rpb1, and rpb2 [Citation16,Citation17]. The results showed that the phylogenetic signals for the five genes were congruent (p = 0.02). Phylogenetic analysis of the five-gene dataset was conducted using maximum-likelihood (ML) methods. The ML analysis was run on RaxML v7.9.1 using the optimal model GTR + I with 1000 rapid bootstrap replicates [Citation18]. The reliability of nodes was assessed 1000 replicates of non-parametric bootstrap proportions on the combined 5-gene dataset.
Table 2. Specimen information for the materials used in this study.
3. Results
3.1. Phylogenetic analysis
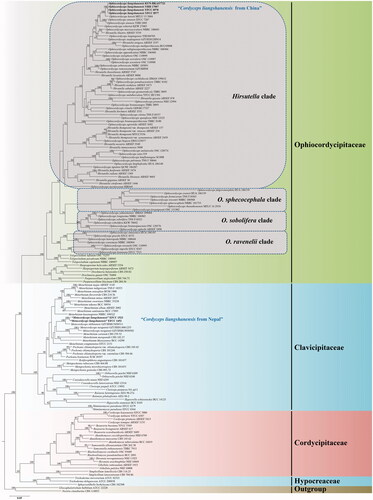
The 149 taxa were used for phylogenetic analysis from four families (Ophicordycipitaceae, Clavicipitaceae, Cordycipitaceae, and Hypocreaceae), with Gliocephalotrichum bulbilium and Nectria cinnabarina included as outgroups. The concatenated sequence dataset of five genes was composed of 5567 bp sequence data (1690 bp for nrSSU, 972 bp for nrLSU, 1035 bp for tef-1α, 781 bp for rpb1, and 1089 bp for rpb2). Phylogenetic tree inferred from ML analysis recognized four statistically well-supported clades in Ophiocordyceps, designated as Hirsutella Pat., O. sobolifera (Hill ex Watson) G.H. Sung et al., O. sphecocephala (Klotzsch ex Berk.) G.H. Sung et al. and O. ravenelii (Berk. & M.A. Curtis) G.H. Sung et al. clades (). The Hirsutella clade included six distinct subclades, namely, H. citriformis Speare, H. thompsonii F.E. Fisher, H. nodulosa Petch, H. Guyana Minter & B.L. Brady, H. sinensis X.J. Liu et al., and the Hirsutella ant pathogen subclades. Phylogenetic analysis of combined dataset placed four samples of C. liangshanensis in the H. sinensis subclade. Cordyceps liangshanensis was closely clustered with O. karstii T.C. Wen, Y.P. Xiao & K.D. Hyde and well-supported by ML bootstrap proportions (ML-BP = 100%). However, three samples of C. liangshanensis clustered together and formed a separate clade from O. karstii with 100% statistical support.
3.2. Morphological features
The morphological characteristics of various specimens of C. liangshanensis are shown, and the photomicrographs of morphological structures are shown in . The detailed fungal morphological descriptions are in the Taxonomy section. Distinct morphological features between C. liangshanensis and its related species are summarized in .
Figure 2. Morphology of Ophiocordyceps liangshanensis. (A) Slender stromata arising from Hepialidae larvae; (B) Mature stroma arising from the larva of Hepialidae. (C) The type specimen of O. liangshanensis (KUN-HKAS7723). (D) The reddish dark brown host of O. liangshanensis. (E) Fertile part; (F–G) Perithecia. (H–J) Asci. (K–M) Ascospores; (N) Colony on PDA; (O–S) Conidiogenous cells; (T–X) Conidia embedded in mucous sheaths. Scale bars: A–C = 3 cm; D, E, N = 1 cm; F = 500 µm; G = 200 µm; H–M = 50 µm; O–S = 10 µm; T–X = 5 µm.
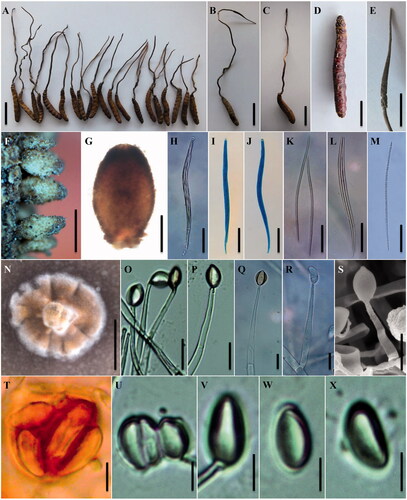
Table 3. Morphological comparisons of Ophiocordyceps liangshanensis with its related species.
3.3. Taxonomy
Ophiocordyceps liangshanensis (M. Zang, D.Q. Liu & R.Y. Hu) H. Yu, Y. Wang, Y.D. Dai, Zhu L. Yang & Y.B. Wang, comb. nov. ().
MycoBank MB837859
≡ Cordyceps liangshanensis M. Zang, D.Q. Liu & R.Y. Hu, Acta Botonic Yunnanica 4(2): 174 (1982).
= Metacordyceps liangshanensis (M. Zang, D. Liu & R.Y. Hu) G.H. Sung, J.M. Sung, Hywel-Jones & Spatafora, Studies in Mycology 57: 35 (2007, misinterpretation).
= Papiliomyces liangshanensis (M. Zang, D. Liu & R.Y. Hu) Luangsa-ard, Samson & Thanakitpipattana, Studies in Mycology 95: 240 (2020, misinterpretation).
Holotype: CHINA. Sichuan Province: Liangshan Yi Autonomous Prefecture, Leibo County, alt. 1500 m, on the larva of Hepialidae sp. living in Qiongzhuea tumidinoda forest, 25 July 1980, Jiyuan Li (KUN-HKAS7723, holotype).
Sexual morph: Stromata cylindrical, solid, yellow-brown to brown, 1–2 arising mainly from the head of host, 200–300 mm long, 1.5–2.5 mm wide. Stipes subcylindrical, slender, and long. Fertile parts cylindrical or clavate, yellow-brown to dark brown, covering apex to middle part of stromata, 30–60 mm long, 2–2.5 mm diam., often with a 3–5 mm long sterile apex (n = 5). Perithecia dense, superficial, long ovoid, with a basal stipe connected to the stromata, becoming yellowish brown to black brown when mature, 450–740 × 300–450 µm (n = 10). Asci hyaline, cylindrical, 8-spored, 260–480 × 8–12 μm (n = 10). Apical caps conspicuous and thick, hemiglobose to taper, 7.2–10.0 µm wide, 4.4–6.4 µm high (n = 10). Ascospores hyaline, fasciculate, thread-like, slender and long, 170–240 × 2.5–4.1 µm (n = 10), with many septa, not breaking into secondary ascospores. Septa, 5.5–19.8 × 2.5–4.1 µm (n = 10).
Asexual morph: Hirsutella. Colonies on PDA growing very slowly, reaching 12–15 mm diam after 2 months at 25 °C, hard, round, irregular swell, brown, and radial growth of white. Cell secretoried dark brown pigment material. Hyphae hyaline, septate, branched, smooth-walled, 3.2–5.4 μm wide (n = 10). Conidiogenous cells monophialidic, sometimes polyphialidic with swollen base and slender neck, generating on hyphae laterally or terminally, hyaline, 46.9–75.6 µm long (n = 20), smooth and subcylindrical in the basal region, reaching 3.8–4.7 µm wide (n = 20), tapering gradually or abruptly to a straight neck, minutely warty, 2.0–3.0 µm wide at the tip (n = 20). Conidia hyaline, aseptate, smooth-walled, arising in groups at the apex of the neck, ellipsoid, citriform or shape of an orange segment, 8.0–12.6 × 3.6–5.0 µm (n = 25), single or 2–4 aggregated, embedded in a pigmented mucous sheath.
Host: Larvae of Hepialidae sp., reddish dark brown, 31–55 mm long, 6–10 mm wide (n = 10).
Other materials examined: CHINA. Sichuan Province: Liangshan Yi Autonomous Prefecture, Leibo County, Xining Town (N 28.26°, E 103.57°), alt. 1540 m, on larvae of Hepialidae sp. living in Qiongzhuea tumidinoda forests, 12 July 2011, Hong Yu (YHH 16800, epitype, designated here; YFCC 8577, ex-epitype living culture); Ibid., 5 August 2016, Lei Ding (YHH 17007–YHH 17050). CHINA. Yunnan Province: Zhaotong City, Shuifu County, Taiping Town, Tongluoba National Forest Park (N 28.41°, E 104.15°), alt. 1750 m, on larvae of Hepialidae sp. living in Q. tumidinoda forests, 20 June 2015, Yong-Dong Dai (YHH 16861–YHH 16900; YFCC 8578, living culture).
Known distribution: this species is distributed in Sichuan, Yunnan, and Guizhou, southwestern China.
4. Discussion
Ophiocordyceps liangshanensis and O. robertsii share similar morphological characteristics by producing long stromata with a sterile apex, wide, and brown perithecia, long and cylindrical asci, except that O. robertsii produces secondary ascospores [Citation19]. There are more than 270 known species of Ophiocordyceps but only a few species (i.e., O. liangshanensis, O. xuefenensis T.C. Wen, R.C. Zhu, J.C. Kang & K.D. Hyde, O. ramosissimum T.C. Wen, J.C. Kang & K.D. Hyde, O. karstii, O. lanpingensis H. Yu & Z. H. Chen, O. emeiensis (A.Y. Liu & Z.Q. Liang) G.H. Sung et al., O. larvarum (Westwood) G.H. Sung et al., and O. sinensis) have long stromata, superficial perithecia and ascospores not breaking into secondary ascospores. Ophiocordyceps liangshanensis differs from the other species mentioned above in having relatively wide perithecia (300–450 µm) (). Its asexual state has long conidiogenous cells, and is similar to that of H. illustris Minter & B.L. Brady and H. strigosa Petch. However, O. liangshanensis differs from H. illustris by its smaller size in conidia (8.0–12.6 × 3.6–5.0 µm). The conidiogenous cells of O. liangshanensis generate on hyaline hyphae laterally or terminally, whereas those of H. strigosa arise at right angle from brown hyphae. To fix the species concept, a recently collected specimen (YHH 16800), is designated here as the epitype of O. liangshanensis.
No serious comparison was made for the materials of C. liangshanensis from China and those from Nepal previously [Citation4,Citation5]. Based on multi-gene phylogeny, “C. liangshanensis” from Nepal reported by Sung et al. [Citation5] was identified as a member in Clavicipitaceae, whereas “C. liangshanensis” from Sichuan (type locality) and Yunnan, China, belongs to Ophiocordycipitaceae in the present study. Thus, its new combination is proposed as Ophiocordyceps liangshanensis instead of the previous names, “Metacordyceps liangshanensis” and “Papiliomyces liangshanensis.”
The genus Papiliomyces, consisting of two species, was proposed for the type species “P. liangshanensis” by Mongkolsamrit et al. [Citation6] based on its phylogenetic placement. However, it is clear that the Nepalese collections differ from the Chinese collections, and should be restudied and described in detail, as well as other nomenclatural and taxonomic confusions should be verified in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Wu P, Wen FY, Wang XY, et al. Investigation and analysis of Cordyceps liangshanensis (in Chinese). Lishizhen Med Mater Med Res. 2019;30(9):2252–2254.
- Liang ZQ, Liu AY, Liu ZY. Cordyceps. In: Flora Fungorum sinicorum, vol. 32. Beijing, China: Science Press; 2007.
- Yang ZL. A study on biology of anamorph of Cordyceps liangshanensis Zang Liu et Hu (in Chinese) [master dissertation]. Kunming, China: Yunnan University; 2011.
- Zang M, Liu DQ, Hu RY. Notes concerning the subdivisions of Cordyceps and a new species from china (in Chinese). Acta Bot Yunnanica. 1982;4(2):173–176.
- Sung GH, Hywel-Jones N, Sung JM, et al. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 2007;57:5–59.
- Mongkolsamrit S, Khonsanit A, Thanakitpipattana D, et al. Revisiting Metarhizium and the description of new species from Thailand. Stud Mycol. 2020;95:171–251.
- Kornerup A, Wanscher JH. Methuen handbook of colour. London, UK: Methuen; 1963.
- Wang YB, Wang Y, Fan Q, et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020;103(1):1–46.
- Wang YB, Yu H, Dai YD, et al. Polycephalomyces agaricus, a new hyperparasite of Ophiocordyceps sp. infecting melolonthid larvae in southwestern China. Mycol Prog. 2015;14:70.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cyptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994;98(6):625–634.
- Bischoff JF, Rehner SA, Humber RA. Metarhizium frigidum sp. nov.: a cryptic species of M. anisopliae and a member of the M. flavoviride complex. Mycologia. 2006;98(5):737–745.
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948.
- Tamura K, Stecher G, Peterson, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Swofford DL. Paup*: Phylogenetic analysis using parsimony (and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates; 2002.
- Lanfear R, Calcott B, Ho SYW, et al. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701.
- Kepler R, Ban S, Nakagir A, et al. The phylogenetic placement of hypocrealean insect pathogens in the genus Polycephalomyces: an application of one fungus one name. Fungal Biol. 2013;117(9):611–622.
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web Servers. Syst Biol. 2008;57(5):758–771.
- Cunningham GH. The genus Cordyceps in New Zealand. Trans N Z Inst. 1921;53:372–382.
- Wen TC, Zhu RC, Kang JC, et al. Ophiocordyceps xuefengensis sp. nov. from larvae of Phassus nodus (Hepialidae) in Hunan Province, southern China. Phytotaxa. 2013;123(1):41–50.
- Wen TC, Xiao YP, Li WJ, et al. Systematic analyses of Ophiocordyceps ramosissimum sp. nov., a new species from a larvae of Hepialidae in China. Phytotaxa. 2014;161(3):227–234.
- Li GJ, Hyde KD, Zhao RL, et al. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78(1):1–237.
- Chen ZH, Dai YD, Yu H, et al. Systematic analyses of Ophiocordyceps lanpingensis sp. nov., a new species of Ophiocordyceps in China. Microbiol Res. 2013;168(8):525–532.
- Liu AY, Liang ZQ, Liu ZY. Cordyceps spp. and some other entomopathogenic fungi from the Emei Mountain Preserve in China (in Chinese). Mycosystema. 1997;16(2):139–143.
- Liang ZQ, Liu AY, Huang JZ, et al. The genus Cordyceps from Kuankuoshui Preserve in Guizhou I (in Chinese). Acta Mycol Sin. 1996;15(4):264–271.