Abstract
Penicillium species are known to be ubiquitous environmental saprophytes. In the survey of diversity of genus Penicillium, seven new records of Penicillium species belonging to section Lanata-Divaricata were isolated from freshwater and soil samples collected from different locations in Korea. Based on morphological characteristics and multilocus phylogenetic analysis of the rDNA internal transcribed spacer region (ITS), β-tubulin (BenA), and calmodulin (CaM) genes, the isolated strains were identified as P. annulatum, P. camponotum, P. echinulonalgiovense, P. globosum, P. limosum, P. onobense, and P. yunnanense, respectively. This study presents detailed phylogenetic analyses and morphological descriptions of these species that contribute to section Lanata-Divaricata in Korea.
1. Introduction
The genus Penicillium established by Link et al. in 1809 [Citation1], has a worldwide distribution, isolated from diverse substrates, including air, soil, freshwater, as endophytes, insect specimens, indoor environments, and food products [Citation2,Citation3]. Using phylogenetic approach, often supported by phenotypic, physiologic and/or extrolite data, members of this genus is divided into two subgenera, 32 sections and 89 series [Citation2]. Members of this genus are economically important as it produce antibiotics, enzymes, organic acids, alcohols and pharmaceuticals [Citation4]. Whereas, some of them cause food spoilage, produce mycotoxins, and cause human and animal diseases [Citation5]. Currently, the genus contains 483 accepted species [Citation2]. In Korea, more than 100 Penicillium species have been reported [Citation6–12].
The Penicillium section Lanata-Divaricata was established by Thom et al. in 1930 [Citation13] for species with biverticillate conidiophores that usually contain an elongation of the conidiophore’s main axis and metulae that diverge from axis to form an asymmetrical verticil. Thus, resulting in conidiophores to be interpreted as monoverticillate, although they are in most cases divergently branched biverticillate conidiophores (also termed divaricate). This group of species mainly isolated from soil, some found from air, and protea repens infructescence [Citation3,Citation14–16]. This section is species-rich, with 56 species are accepted until 2016 [Citation14–19]. But the list is rapidly increasing with many new Penicillium species recently described from all over the world and added to section Lanata-Divaricata. Up to date, 76 species of Penicillium sect. Lanata-Divaricata have been accepted [Citation2]. Recently, P. soli, P. melanosporum, P. siccitolerans and P. michoacanense, were discovered from phosphate solubilizing soil in China and soil samples as xerophilic in Mexico and Spain [Citation20,Citation21].
To our knowledge, only 16 species of Penicillium section Lanata-Divaricata have been reported in Korea until now [Citation6,Citation10,Citation11]. Thus, the aim of this study was to isolate, identify and describe the previously unrecorded seven isolates found in soil and freshwater samples collected from different locations in Korea, P. annulatum, P. camponotum, P. echinulonalgiovense, P. globosum, P. limosum, P. onobense, and P. yunnanense based on multi-loci phylogenetic analysis of ITS, BenA and CaM, and morphological data.
2. Materials and methods
2.1. Sampling and isolation
Details of freshwater and soil samples collected from different locations in South Korea are shown in . Serial dilution plating method was used to isolate fungal strains; 1 g of soil or 1 mL of freshwater sample was added to 9 mL sterile distilled water. Approximately, 10−1, 10−2, 10−3, and 10−4 dilutions were plated onto potato dextrose agar (PDA) (DifcoTM, Sparks, MD, USA) and malt extract agar (MEA) (DifcoTM) containing 50 ppm streptomycin solution. Plates were incubated at 25 °C for 7–10 days. Then, colonies were transferred to new PDA plates and incubated for 7 days at 25 °C.
Table 1. Information of Penicillium isolates used in this study.
For stock storage, pure isolates were maintained in 20% glycerol at −80 °C and in PDA slant tubes in the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea. The isolated strains were also deposited in the Collection of National Institute of Biological Resources (NIBR), Incheon, and Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea as shown in .
2.2. DNA extraction and PCR sequencing
Total genomic DNA was extracted directly from the mycelia using Solg TM Genomic DNA Prep Kit (Solgent Co. Ltd., Daejeon, Korea). The ITS rDNA region was amplified with primer pairs ITS1/ITS4 or ITS5/LR5 [Citation22], BenA gene with T10/Bt2b [Citation23] or T1/Bt2b [Citation23,Citation24], and CaM gene were amplified with CF1/CF4 [Citation25], respectively. The PCR cycling programs used for amplification follows: initial denaturation at 94 °C for 4 min, followed by denaturing at 94 °C for 1 min, then annealing (in case of ITS rDNA and BenA) at 56 °C for 30 s (in case of CaM, annealing temperature at 55 °C for 50 s), extension for 2 min at 72 °C, and a final 10 min elongation step at 72 °C which was followed by cooling at 4 °C for 30 cycles. PCR products were visualized in 1% (w/v) agarose gel electrophoresis. The PCR products were purified with the Accuprep PCR Purification Kit (Bioneer Corp., Daejeon, Korea). PCR products were sequenced using the same PCR primers on an ABI PRISM3730XL Genetic Analyzer (Applied Biosystems, CA, USA) at Macrogen (Daejeon, Korea).
2.3. Phylogenetic analysis
All sequence data used in this study were obtained from GenBank. Sequences were aligned with Clustal_X version 2.1 [Citation26] and were edited manually with Bioedit version 7.2.6.0 [Citation27]. Maximum likelihood (ML) phylogenies were assessed using MEGA 7 software [Citation28] and the Kimura 2-parameter model. The p-distance substitution model with 1,000 bootstrap replications was used for the assessment of the reliability of internal branches. Penicillium glabrum CBS 125543 (T) was used as outgroup. The sequences of the isolates in this study were deposited in the database under the accession numbers shown in .
Table 2. GenBank accession numbers for fungal strains used in this study.
2.4. Morphological studies
For characterization, the respective strains were inoculated into three points, namely Czapek yeast autolysate agar (CYA), Blakeslee’s malt extract agar (MEA), and yeast extract sucrose agar (YES), and incubated at 25 °C for 7 days [Citation29]. For morphological observations, fragments of mycelia were removed from the cultures and placed on microscope slides with lactic acid (60%). An Olympus BX51 microscope (Olympus, Tokyo, Japan) was used to capture digital images.
3. Results
3.1. Phylogenetic analysis
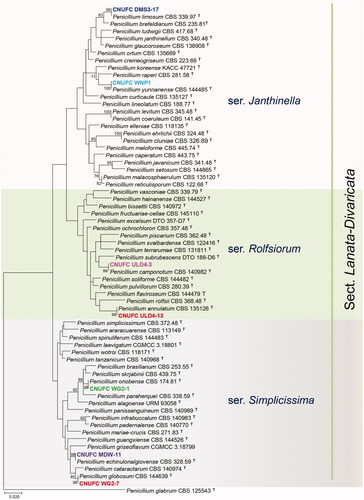
A BLASTn search on the ITS, BenA, and CaM regions of the isolates were obtained. ITS regions of CNUFC ULD4-13, CNUFC ULD4-3, CNUFC MDW-11, CNUFC WG2-7, CNUFC DMS3-17, CNUFC WG2-1, and CNUFC WNP1 showed similarities of 99.8% (498/499 bp), 99.8% (485/486 bp), 99.8% (501/502 bp), 99.8% (488/489 bp), 100% (533/533 bp), 100% (533/533 bp), and 99.8% (481/482 bp) with P. annulatum (NR_138303), P. camponotum (NR_158823), P. echinulonalgiovense (GU981587), P. globosum (KY495018), P. limosum (NR_111496), P. onobense (NR_111497), and P. yunnanense (KY494989), respectively. In a BLASTn search with BenA sequence, the isolate CNUFC ULD4-13, CNUFC ULD4-3, CNUFC MDW-11, CNUFC WG2-7, CNUFC DMS3-17, CNUFC WG2-1, and CNUFC WNP1 showed similarity 99.6% (470/472 bp), 99.17% (479/483 bp), 98.80% (411/416 bp), 100% (416/416 bp), 99.79% (466/467 bp), 100% (480/480 bp), and 99.76% (411/412 bp) with sequences of P. annulatum CV0037 (JX091514), P. camponotum (KT887819), Penicillium sp. YD-2017a (KY495150), P. globosum (KY495127), P. limosum (GU981621), P. onobense (GU981627), and P. yunnanense (KY495099), respectively. Similarly, CaM sequence of CNUFC ULD4-13, CNUFC ULD4-3, CNUFC MDW-11, CNUFC WG2-7, CNUFC DMS3-17, CNUFC WG2-1, and CNUFC WNP1 showed similarities of 98.53% (536/544 bp), 98.64% (436/442 bp), 98.51% (398/404 bp), 99.5% (402/404 bp), 100% (407/407 bp), 99.75% (405/406 bp), and 99.75% (405/406 bp) with P. annulatum (JX141547), P. camponotum (KT887781), Penicillium sp. YD-2017a (KY494981), P. globosum (KY494958), P. limosum (KF296398), P. onobense (KF296371), and P. yunnanense (KY494930), respectively. Phylogenetic tree based on the combined sequence data of the three loci, ITS, BenA, and CaM revealed that the seven isolated strains were identical to P. annulatum, P. camponotum, P. echinulonalgiovense, P. globosum, P. onobense, P. limosum, and P. yunnanense ().
Figure 1. Phylogenetic tree of Penicillium annulatum CNUFC ULD4-13, P. camponotum CNUFC ULD4-3, P. echinulonalgiovense CNUFC MDW-11, P. globosum CNUFC WG2-7, P. limosum CNUFC DMS3-17, P. onobense CNUFC WG2-1, and P. yunnanense CNUFC WNP1 and related species belonging to section Lanata-Divaricata based on maximum likelihood analysis of the combined ITS, BenA, and CaM sequences. Numbers at the nodes indicate the bootstrap values (>70%) from 1,000 replicates. The bar indicates the number of substitutions per nucleotide. The study isolates are shown in bold and different colors.
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC ULD4-13
Penicillium annulatum Visagie & K. Jacobs, Mycological Progress 14 (10/96): 14 (2015) ().
Figure 2. Morphology of Penicillium annulatum. (A, D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
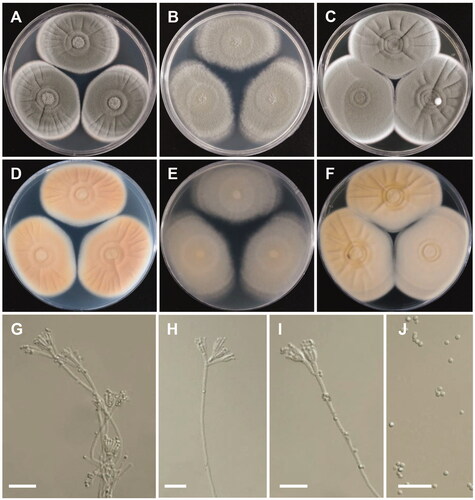
Colony characteristics: CYA 25 °C, 7 days: colonies were moderately deep, radially and concentrically sulcate, with ring-like appearance because of sporulating and nonsporulating areas, with low margins, light-green to white mycelia, floccose texture, sparse to moderately dense sporulation, no soluble pigment, reverse light to grayish orange, and reached 40–46 mm in diameter. MEA 25 °C, 7 days: colonies were low to moderately deep, plane, low, irregular margins, light-green to white mycelia, floccose texture, moderately dense to dense sporulation, no exudate, no soluble pigment, reverse dull-green to greenish white, and reached 42–47 mm in diameter. YES 25 °C, 7 days: colonies were radially sulcate, with ring-like appearance because of sporulating and nonsporulating areas but less than those on CYA, low, narrow margins, white mycelia, floccose texture, sparse to moderate sporulation, no exudate and soluble pigment, reverse dull-yellow to yellowish white, and reached 41–49 mm in diameter.
Micromorphology: Conidiophores were bi- and terverticillate, 165–720 × 3–4.5 µm. Metulae were appressed to divergent, 7.8–20 × 2.5–4.5 µm. Phialides were ampulliform 3–5 per metula, 6.2–8 × 2–3.5 µm. Conidia were globose to subglobose, 2.5–3 × 2–3 µm.
3.2.2. Taxonomy of CNUFC ULD4-3
Penicillium camponotum Visagie, David Clark, & Seifert, Persoonia 36: 271 (2016) ().
Figure 3. Morphology of Penicillium camponotum. (A,D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
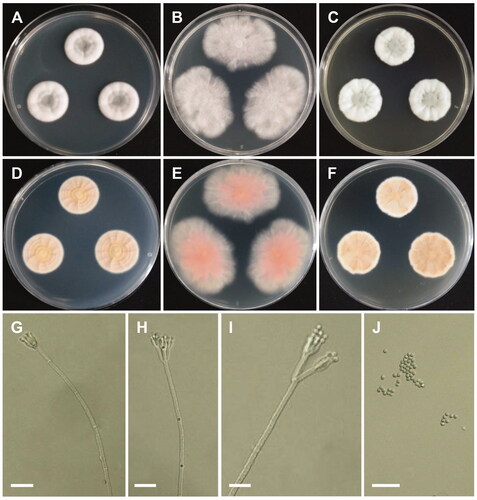
Colony characteristics: CYA 25 °C, 7 days: colonies were moderately deep, lightly radially sulcate, narrow, entire margins, white mycelia, floccose texture, sparse to absent sporulation, no soluble pigments, reverse yellowish white to pale-yellow, and reached 20–22 mm in diameter. MEA 25 °C, 7 days: colonies were plane, entire margins, white mycelia, floccose texture; moderately dense sporulation, no soluble pigments, no exudates, reverse pinkish gray, and reached 21–26 mm in diameter. YES 25 °C, 7 days: colonies were moderately deep, radially, and concentrically sulcate, with low, wide, entire margins, white mycelia, floccose texture, moderately dense sporulation, no soluble pigments, reverse light-brown, and reached 21–23 mm in diameter.
Micromorphology: Conidiophores were biverticillate, 212–610 × 2.5–3.5 µm. Metulae were divergent, 12.2–25.6 × 2.5–4 µm. Phialides were ampulliform, 3–6 per metula, 8.2–11.4 × 2.5–3.5 µm. Conidia were globose, 2.2–3.5 × 2.1–3.4 µm.
3.2.3. Taxonomy of CNUFC MDW-11
Penicillium echinulonalgiovense S. Abe ex Houbraken & R.N. Barbosa, Antonie van Leeuwenhoek 111 (10): 1895 (2018) ().
Figure 4. Morphology of Penicillium echinulonalgiovense. (A,D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
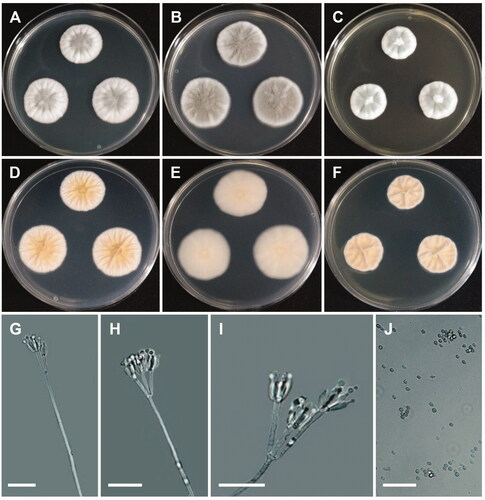
Colony characteristics: CYA 25 °C, 7 days: colonies were moderately deep and lightly radially sulcate, with low, narrow, entire margins, white mycelia, floccose texture, sparse to absent sporulation, no soluble pigments, reverse yellowish white to pale-yellow, and reached 18–22 mm in diameter. MEA 25 °C, 7 days: colonies were low to moderately deep and plane, with low, irregular margins, light-green to white mycelia, floccose texture, moderately dense to dense sporulation, no exudate, no soluble pigment, reverse dull-green to greenish white, and reached 20–23 mm in diameter. YES 25 °C, 7 days: colonies were low to moderately deep, sunken at center, radially and concentrically sulcate, with low, narrow, entire margins, white mycelia, floccose texture, sparse to moderately dense sporulation, no exudate and soluble pigment, reverse dull-green, and reached 17–20 mm in diameter.
Micromorphology: Conidiophores were biverticillate, 56–210 × 2.2–3.0 µm. Metulae were divergent, 10.7–18.8 × 2.2–3.1 µm. Phialides were ampulliform, 2–6 per metula, 7.1–10.9 × 2.5–3.0 µm. Conidia were echinulate, globose to subglobose 2.5–3.9 × 2.3–3.7 µm.
3.2.4. Taxonomy of CNUFC WG2-7
Penicillium globosum L. Cai, Houbraken, & X.Z. Jiang, Cladistics 35 (5): 529 (2018) ().
Figure 5. Morphology of Penicillium globosum. (A,D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
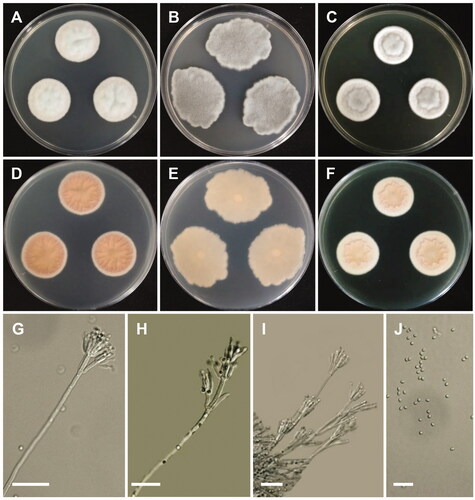
Colony characteristics: CYA 25 °C, 7 days: colonies were sunken in center, radially and concentrically sulcate, low, narrow, entire margins, blueish white mycelia, floccose texture, sparse to moderately dense sporulation, no soluble pigment, reverse pale-orange to apricot, and reached 20–22 mm in diameter. MEA 25 °C, 7 days: colonies were low, irregular margins, white mycelia, floccose texture, sparse to moderately dense sporulation, no exudate, no soluble pigment, reverse grayish green, and reached 20–24 mm in diameter. YES 25 °C, 7 days: colonies were deep, raised at center, radially and concentrically sulcate, low, narrow, entire margins, white mycelia, floccose, sulcate texture, sparse to moderately dense sporulation, no exudate, no soluble pigment, reverse grayish orange, and reached 16–19 mm in diameter.
Micromorphology: Conidiophores were mono- and biverticillate, 42–162 × 3.6–4 µm. Metulae were appressed to divergent, 7.6–20.2 × 2.7–4 µm. Phialides were ampulliform, 3–9 per metula, 7.8–12.6 × 2.2–4 µm. Conidia were globose to subglobose, 2.9–4.1 × 2.1–3.8 µm.
3.2.5. Taxonomy of CNUFC DMS3-17
Penicillium limosum S. Ueda, Mycoscience 36 (4): 451 (1995) ().
Figure 6. Morphology of Penicillium limosum. (A,D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–J) Conidiophores; (K) Conidia (scale bars: G–K = 20 μm).
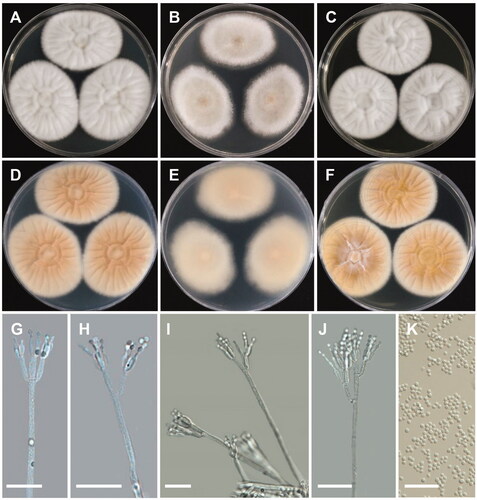
Colony characteristics: CYA 25 °C, 7 days: colonies were radially sulcate at center, with light grayish green to white floccose surface, no exudate and soluble pigment, reverse brown to brownish orange, and reached 39–44 mm in diameter. MEA 25 °C, 7 days: colonies were plain, white to grayish green mycelia, with sparse to moderately dense sporulation, no soluble pigment, reverse grayish yellow, and reached 35–41 mm in diameter. YES 25 °C, 7 days: colonies were deep, raised at center, radially and concentrically sulcate, with low, narrow, entire margins, white mycelia, floccose, sulcate texture, sparse to moderately dense sporulation, no exudate, no soluble pigment, reverse grayish orange, and reached 36–40 mm in diameter.
Micromorphology: Conidiophores were biverticillate, 73–252 µm. Metulae were appressed to divergent, 14.8–20.2 × 3–4 µm. Phialides were ampulliform, 5–6 per metula, 8–12.6 × 2.2–3 µm. Conidia were globose to subglobose, 2.7–3.2 × 2.4–3 µm.
3.2.6. Taxonomy of CNUFC WG2-1
Penicillium onobense C. Ramírez & A.T. Martínez, Mycopathologia 74 (1): 44 (1981) ().
Figure 7. Morphology of Penicillium onobense. (A,D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
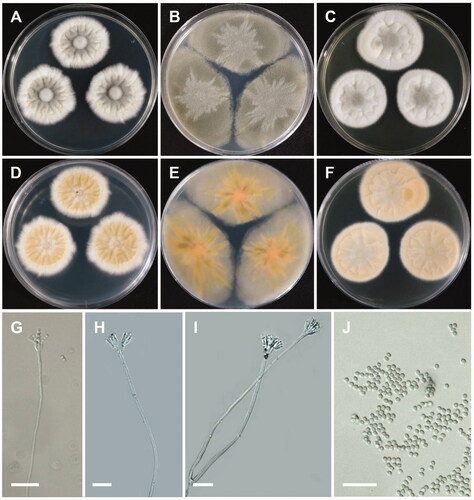
Colony characteristics: CYA 25 °C, 7 days: colonies were velvety, radiately wrinkled, dirty white, entire margins, white mycelia, velutinous texture, moderately dense sporulation, no soluble pigments and exudates, reverse pale-brownish orange, and reached 19–22 mm in diameter. MEA 25 °C, 7 days: colonies were low, with low, irregular margins, green mycelia, grayish green at center, floccose texture, sparse to moderately dense sporulation, no exudate, no soluble pigment, reverse grayish green with light orange circle at center, and reached 36–43 mm in diameter. YES 25 °C, 7 days: colonies were moderately deep, radially and concentrically sulcate, with low, wide, entire margins, white mycelia, floccose texture, moderately dense sporulation, no soluble pigments, no exudates, reverse light-brown, and reached 18–23 mm in diameter.
Micromorphology: Conidiophores were biverticillate, 73–310 µm. Metulae were divergent, 10.2–13.1 × 3.3–5 µm. Phialides were ampulliform, 3–6 per metula, 8.3–12.2 × 2.2–3 µm. Conidia were elliptical to subglobose, 2.3–4.1 × 2.1–4.1 µm.
3.2.7. Taxonomy of CNUFC WNP1-1
Penicillium yunnanense L. Cai & X.Z. Jiang, Cladistics 35 (5): 545 (2018) ().
Figure 8. Morphology of Penicillium yunnanense. (A,D) Colonies on Czapek yeast autolysate agar (CYA); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on yeast malt extract agar (YES). (A–C: obverse view, D–F: reverse view). (G–J) Conidiophores; (K) Conidia (scale bars: G–K = 20 μm).
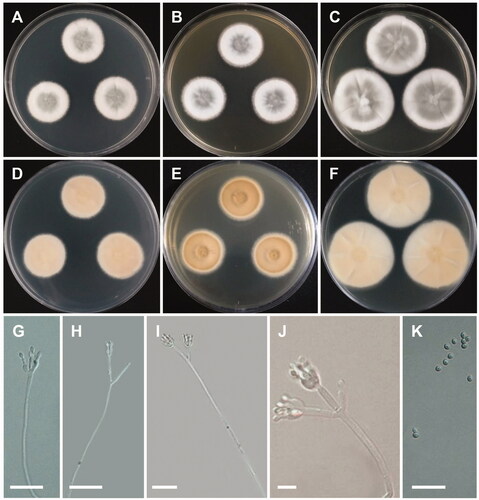
Colony characteristics: CYA 25 °C, 7 days: colonies were moderately deep, raised at center, radially sulcate, with low, narrow, entire margins, white mycelia, velutinous texture, moderately dense sporulation, no soluble pigments, no exudates, reverse greenish gray to dull-green, and reached 22–24 mm in diameter. MEA 25 °C, 7 days: colonies were moderately deep, radially sulcate, with low, narrow, irregular margins, white mycelia, floccose texture, sparse to moderately dense sporulation, no exudate, no soluble pigment, reverse pale to yellowish gray, pale yellowish circles at center, and reached 23–24 mm in diameter. YES 25 °C, 7 days: colonies were low to moderately deep, sunken at center, radially and concentrically sulcate, with low, narrow, entire margins, white mycelia, floccose texture, sparse to moderately dense sporulation, no exudate, no soluble pigment, reverse dull-green, and reached 26–28 mm in diameter.
Micromorphology: Conidiophores were mono- and biverticillate, 22–126 × 2–3 µm. No metulae were observed. Phialides were ampulliform, 6.6–14.1 × 2–4 µm. Conidia were broadly ellipsoidal to ellipsoidal, 3–4 × 2.5–3.5 µm.
4. Discussion
This study advanced our understanding of Penicillium sect. Lanata-Divaricata and contributed seven new records for Korea.
Although ITS rDNA is designated as the universal primers available and most widely sequenced marker for fungi [Citation30], but not variable enough for distinguishing all closely related species in Penicillium [Citation3,Citation31]. Thus, additional secondary marker such as BenA and CaM were proposed and found to be useful for the accurate identification of Penicillium species [Citation2,Citation3,Citation15]. Phylogenetic tree based on combined ITS-BenA-CaM sequences indicated that seven strains belonging to the series Janthinella, Rolfsiorum and Simplicissima in section Lanata-Divaricata ().
The isolates CNUFC ULD4-13 was well placed within the clade of P. annulatum in series Rolfsiorum (). The isolate was morphologically similar to description of P. annulatum [Citation15] with respect to producing bi- and terverticillate conidiophores, ampuiliform phialides, and roughened globose to subglobose conidia (). Previous studies showed P. annulatum to be isolated from air sample, soil, and mite in Protea repens infructescence and Stellenbosch in South Africa [Citation15]. There are no studies on extrolites produced by P. annulatum. Similarly, the phylogenetic placement and morphological characteristics of isolates CNUFC ULD4-3 into series Rolfsiorum (), matched previously with described species, P. camponotum [Citation16]. Penicillium camponotum isolated from carpenter ants from New Brunswick, Canada, and ant nest in Picea abies from Germany, was reported to produce andrastin A, B, and C, citrinalin, mangrovamides, marcfortine A and B, and patulin [Citation16]. In this study, P. camponotum was isolated from soil samples. The isolate CNUFC MDW-11 was grouped with P. echinulonalgiovense in series Simplicissima () and shared morphology similar to the previous descriptions. Penicillium echinulonalgiovense was first isolated from soil samples in Japan without a Latin diagnosis, later validated by Barbosa et al. [Citation32,Citation33]. Penicillium echinulonalgiovense was also isolated from bee pollen and nest of Melipona scutellaris located at Recife, Pernambuco in Brazil; soil samples from Australia, China, Hong Kong, Indonesia, USA, Madagascar, and Malaysia; and industrial installations in Netherlands [Citation33]. Penicillium echinulonalgiovense produces andrastin A, xanthoepocin, and pulvilloric acid [Citation2,Citation33]. Similarly, CNUFC WG2-7 was clustered within the same clade as P. globosum in series Simplicissima (), described by Diao et al. [Citation34]. The isolate CNUFC WG2-7 was morphologically similar to P. globosum with only differences in the number of phialides per metula. According to Diao et al. [Citation34], 3–19 phialides per metula whereas the isolate CNUFC WG2-7 consists of 3–9 phialides per metula. P. globosum was isolated from an acidic soil in China and Australia; rainforest soil from Malaysia; industrial installations in Netherlands; and soil in citrus grove in Florida, USA [Citation34]. Also, the morphological characteristics of CNUFC DMS3-17 were similar to the previous descriptions [Citation35] and was also clustered in P. limosum CBS 339.97 (type) in series Janthinella (). Earlier, Penicillium limosum have been reported to be found only in marine sediment in Nagasaki Prefecture, Japan [Citation35]. In this study, we isolated P. limosum from damp soil samples. Penicillium limosum was reported to produce a sexual state [Citation35] but no sexual state is found in CNUFC DMS3-17 isolate. Based on previous descriptions, CNUFC WG2-1 shared similar morphology and placed in series Simplicissima (). Penicillium onobense isolated from soil and andosol in Navarra, Spain [Citation36], produces brefeldin A, janthitrems/shearinins, and 2-(4-hydroxyphenyl)-2-oxoacetaldehyde oxime [Citation2]. The isolate CNUFC WNP1 was placed in series Janthinella () and also shared similar morphological characters with previous descriptions [Citation34]. P. yunnanense was isolated from acidic soil in China [Citation34]. Interestingly, this study isolated P. echinulonalgiovense, P. globosum, P. onobense, and P. yunnanense from freshwater for the first time.
This study on the isolation and description of seven new records of Penicillium sect. Lanata-Divaricata from freshwater and soil samples adds to our knowledge on fungal biodiversity. Different environmental sources such as bees, ants, flowers, leaves, nut kernels, and shells studies are needed in Korea considering the increasing evidence for ecological specialization in Penicillium species. Additional studies on the production of extracellular enzymes, antimicrobial compounds, and extrolites are still needed in the genus Penicillium.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Link HF. Observationes in ordines plantarum naturales. Dissertatio 1. Mag Ges Naturf Freunde Berlin. 1809;3:3–42.
- Houbraken J, Kocsubé S, Visagie CM, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169.
- Visagie CM, Houbraken J, Frisvad JC, et al. Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014;78:343–371.
- Frisvad JC, Smedsgaard J, Larson TO, et al. Mycotoxin, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004;49:201–241.
- Pitt JI. The current role of Aspergillus and Penicillium in human and animal health. Med Mycol. 1994;32(s1):17–32.
- Kim HJ, Kim JS, Cheon KH, et al. Species list of Aspergillus, Penicillium and Talaromyces in Korea, based on ‘One Fungus One Name’ System. Kor J Mycol. 2016;44:207–219.
- Pangging M, Nguyen TTT, Lee HB. New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology. 2019;47(2):154–164.
- Nguyen TTT, Pangging M, Bangash NK, et al. Five new records of the family Aspergillaceae in Korea, Aspergillus europaeus, A. pragensis, A. tennesseensis, Penicillium fluviserpens, and P. scabrosum. Mycobiology. 2020;48(2):81–94.
- You YH, Cho HS, Song J, et al. Penicillium koreense sp. nov., isolated from various soils in Korea. J Microbiol Biotechnol. 2014;24(12):1606–1608.
- Park SP, Lee JW, Kim SH, et al. Penicillium from rhizosphere soil in terrestrial and coastal environments in South Korea. Mycobiology. 2020;48(6):431–442.
- Choi DH, Kim YG, Lee IS, et al. Identification and characterization of unreported Penicillium species in Korea. Kor J Mycol. 2020;48:445–456.
- Choi DH, You YH, Lee IS, et al. Penicillium ulleungdoense sp. nov. from Ulleung Island in Korea. Mycobiology. 2020;49(1):46–53.
- Thom C. The penicillia. Baltimore: Williams and Wilkins Co.; 1930.
- Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70(1):1–51.
- Visagie CM, Houbraken J, Seifert KA, et al. Four new Penicillium species isolated from the fynbos biome in South Africa, including a multigene phylogeny of section Lanata-Divaricata. Mycol Prog. 2015;14:96–119.
- Visagie CM, Renaud JB, Burgess KMN, et al. Fifteen new species of Penicillium. Persoonia. 2016;36:247–280.
- Taniwaki MH, Pitt JI, Iamanaka BT, et al. Penicillium excelsum sp. nov from the Brazil nut tree ecosystem in the Amazon Basin'. PLoS One. 2015;10(12):e0143189.
- Laich F, Andrade J. Penicillium pedernalense sp. nov., isolated from whiteleg shrimp heads waste compost. Int J Syst Evol Microbiol. 2016;66(11):4382–4388.
- Zhou QX, Houbraken J, Li QR, et al. Diversity of Penicillium species isolated from heavy metal polluted soil in Guizhou Province. China. Phytotaxa. 2016;273(3):167–174.
- Doilom M, Guo JW, Phookamsak R, et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front Microbiol. 2020;11:585215.
- Rodríguez-Andrade E, Stchigel AM, Cano-Lira JF. New xerophilic species of Penicillium from soil. JoF. 2021;7(2):126.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 315–322.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116.
- Peterson SW, Vega FE, Posada F, et al. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97(3):659–666.
- Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Samson RA, Houbraken J, Thrane U, et al. Food and indoor fungi. CBS laboratory manual series 2. Utrecht: CBS KNAW Fungal Biodiversity Centre. 2010. p. 390.
- Schoch CL, Seifert KA, Huhndorf S, et al.; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246.
- Seifert KA, Samson RA, deWaard JR, et al. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci USA. 2007;104(10):3901–3906.
- Abe S. Studies on the classification of the Penicilla. J Gen Appl Microbiol. 1956;2(1-2):1–344.
- Barbosa RN, Bezerra JDP, Souza-Motta CM, et al. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek. 2018;111(10):1883–1912.
- Diao YZ, Chen Q, Jiang XZ, et al. Penicillium section Lanata-divaricata from acidic soil. Cladistics. 2019;35(5):514–549.
- Ueda S. A new species of Eupenicillium from marine sediment. Mycoscience. 1995;36(4):451–454.
- Ramírez C, Martínez AT. Seven new species of Penicillium and a new variety of Penicillium novae-caledoniae Smith. Mycopathogia. 1981;74:44.
