Abstract
The Federated States of Micronesia (FSM) is an island country in the western Pacific and is a known biodiversity hotspot. However, a relatively small number of fungi (236 species) have been reported till July 2021. Since fungi play major ecological roles in ecosystems, we investigated the fungal diversity of FSM from various sources over 2016 and 2017 and constructed a local fungal inventory, which also included the previously reported species. Fruiting bodies were collected from various host trees and fungal strains were isolated from marine and terrestrial environments. A total of 99 species, of which 78 were newly reported in the FSM, were identified at the species level using a combination of molecular and morphological approaches. Many fungal species were specific to the environment, host, or source. Upon construction of the fungal inventory, 314 species were confirmed to reside in the FSM. This inventory will serve as an important basis for monitoring fungal diversity and identifying novel biological resources in FSM.
1. Introduction
The Federated States of Micronesia (FSM) is an island country in the western Pacific. It consists of four states (from west to east): Yap, Chuuk, Phohnpei, and Kosrae. The climate is warm and humid-tropical rainforest throughout the year, and the landscape of each state varies from low coral atolls to high mountainous terrain. The FSM is a biodiversity hotspot, with 3025 species of animals and 1553 species of plants reported hitherto [Citation1]. However, a relatively small number of fungi (236 species) have been reported [Citation1–19]. The magnitude of fungal species is estimated conservatively to be 1.5 M [Citation20]. Given that the fungus to plant ratio (6:1) is one of the key elements for estimating the number of fungal species, the fungal species reported in the FSM is extremely low.
Fungi play major ecological roles as saprotrophs, symbionts, and pathogens in various environments [Citation21]. Since the FSM comprises various types of habitats, its fungal species is believed to be more diverse than previously reported. Studies on fungal diversity in the FSM were limited to polypores in the forests [Citation7], indoor fungi [Citation16,Citation17,Citation19], and plant pathogens [Citation2,Citation10]. Previous studies on fungal identification in the FSM were mostly confined to the comparisons of morphological characteristics and seldom on sequence analysis [Citation16,Citation17,Citation19]. Therefore, limited fungal sequence data from the FSM are available.
As part of the projects organized by the Ministry of Ocean and Fisheries in Korea, we investigated fungal diversity from various sources in the FSM in 2016 and 2017. The aim of this study was to survey the fungi in the FSM using combined molecular and morphological approaches and construct a new fungal inventory based on the species identified in this study as well as those from previously reported studies.
2. Materials and methods
2.1. Sample collection and isolation
Fungal sampling was performed in the states of Chuuk (January 2016) and Kosrae (January 2017) in the FSM (). Fruiting bodies were collected from the forest, while fungal strains were isolated from various sources in both terrestrial (rotten woods) and marine environments (sea sand and seaweeds). Sampling and transport processes were performed with the permission of the FSM Government.
The fruiting bodies were dried completely and transported to the laboratory in Korea. The specimens were initially grouped based on their morphological features [Citation22–27]. Representative specimens (between 1–3) from each group were selected for molecular identification. Specimens were also deposited at the Seoul National University Fungus Collection (SFC) ().
Table 1. List of species identified from the fruiting bodies collected from Chuuk and Kosrae, the Federated States of Micronesia.
To isolate the fungal strain, 5 g of sea sand was mixed with ten times its volume of sterile water, and 100 μL of the supernatant was plated on Dichloran Rose Bengal Chloramphenicol agar (DRBC; Difco, Becton Dickinson). The rotten wood and seaweed were cut to approximately 5 mm in length and plated on DRBC agar. The plates were transported to the laboratory in Korea in an icebox. All plates were incubated at 25 °C for 5–7 d. Individual fungal strains were transferred to potato dextrose agar (PDA; Difco, Becton Dickinson) plates and stored in 20% glycerol at −80 °C at the SFC (). Fungal strains were grouped morphologically according to their respective genera.
Table 2. List of species identified from the fungal strains isolated from different sources in Chuuk and Kosrae, the Federated States of Micronesia.
2.2. Molecular identification processes
Genomic DNA was extracted from the fruiting bodies and fungal strains using a modified cetyltrimethylammonium bromide extraction protocol [Citation28]. PCR amplification of the internal transcribed spacer (ITS) of all fruiting bodies and fungal strains was performed using the primers ITS1F and ITS4 [Citation29,Citation30]. Appropriate protein-coding genes were used for the accurate identification of some genera; calmodulin (CaM) for Aspergillus, β-tubulin (benA) for Penicillium, and translation elongation factor 1-α (tef1) for Fusarium and Trichoderma were amplified using the primers CF1 and CF4 [Citation31], Bt2a and Bt2b [Citation32], and EF1 and EF2 [Citation33], respectively. All PCRs were performed using AccuPower® PCR premix (Bioneer Co., Daejeon, Korea) in a C1000 thermal cycler (Bio-Rad, Richmond, CA). The PCR conditions for all loci were as follows: initial denaturation for 5 min at 95 °C followed by 35 cycles of 40 s at 95 °C, 40 s at 55 °C, and 60 s at 72 °C, with a final extension for 10 min at 72 °C. The PCR products were purified using the ExpinTM PCR Purification Kit (GeneAll Biotechnology, Seoul, Korea). DNA sequencing was conducted using an ABI Prism 3700 Genetic Analyzer (Life Technologies, Gaithersburg, MD, USA) at Macrogen (Seoul, Korea).
The sequences were assembled, proofread, and edited using MEGA5 [Citation34]. The revised sequences were uploaded to GenBank and the accession numbers are listed in Supplementary Table 1. Genus-by-genus phylogenetic analyses were performed for molecular identification. Multiple generic sequence alignments were performed using MAFFT v7 [Citation35]. Neighbor-joining trees were constructed with MEGA5 using the Kimura 2-parameter model [Citation36] with 1000 bootstrap replicates.
2.3. Species inventory
A list of previously reported fungal species in the FSM was compiled using the occurrence data from the Global Biodiversity Information Facility [Citation1] (https://www.gbif.org/occurrence/download/0204233-200613084148143), New Zealand Fungal Herbarium [Citation2], and other publications (Table S1). The species newly reported in this study were also included in this fungal inventory.
3. Results
3.1. Species identification
A total of 118 fruiting bodies were collected from Chuuk (n = 56) and Kosrae (n = 62). Based on the morphological features and phylogenetic analysis of ITS sequences, 46 different species (two phyla, nine orders, 22 families, and 38 genera) were identified (). Among the 118 fruiting bodies, 107 (90.7%) belonged to Basidiomycota and 11 (9.3%) belonged to Ascomycota (). The most dominant order was Polyporales (50.0%, n = 59), followed by Agaricales (25.4%, n = 30), Auriculariales (7.6%, n = 9), and Xylariales (7.6%, n = 9) (). The dominant species were Leiotrametes menziesii (n = 11), followed by Auricularia polytricha (n = 9), Fomitopsis ostreiformis (n = 7), Favolus grammocephalus (n = 6), and Schizophyllum commune (n = 6) (, , )).
Figure 2. Composition of fruiting bodies (A) and fungal strains (B) from Chuuk and Kosrae, the Federated States of Micronesia.
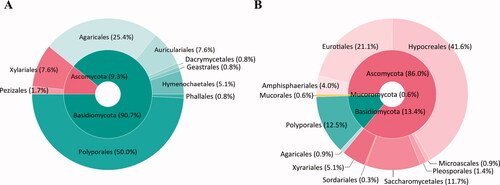
Figure 3. Major species of fruiting bodies (A–H) and fungal strains (I-P) from Chuuk and Kosrae, the Federated States of Micronesia. (A) Auricularia polytricha, (B) Coprinellus disseminates, (C) Fomitopsis ostreiformis, (D) Funalia aspera, (E) Leiotrametes menziesii, (F) Lentinus squarrosulus, (G) Marasmius palmivorus, (H) Tinctoporellus epimiltinus, (I) Aspergillus aculeatinus, (J) Aspergillus brunneoviolaceus, (K) Aspergillus neoniger, (L) Galactomyces candidum, (M) Gongronella butleri, (N) Penicillium citrinum, (O) Trichoderma guizhouense, and (P) Trichoderma reesei.

A total of 351 fungal strains were isolated from Chuuk (n = 325) and Kosrae (n = 26), of which 58 species (three phyla, 11 orders, 21 families, and 28 genera) were identified based on genus-by-genus phylogenetic analyses of ITS, act, benA, CaM, and tef1 (). Five out of the 58 species were also obtained as fruiting bodies. Of the 351 strains, 302 (86.0%) belonged to Ascomycota, 47 (13.4%) belonged to Basidiomycota, and two (0.6%) belonged to Mucoromycota (). Hypocreales (41.6%, n = 146), Eurotiales (21.1%, n = 74), and Polyporales (12.5%, n = 44) were the dominant orders (). The dominant species were Trichoderma reesei (n = 115), followed by Galactomyces candidum (n = 41), F. ostreiformis (n = 37), and Penicillium citrinum (n = 32) (, , )).
3.2. Source comparison
Fruiting bodies were collected from various host trees: Mangifera indica (19 fungal species from 21 strains), Cocos nucifera (13 spp. from 19 strains), Artocarpus altilis (12 spp. from 14 strains), Rhizophora sp. (9 spp. from 14 strains), and others (31 spp. from 50 strains) (). L. menziesii and A. polytricha were commonly found across various host trees, whereas many other species were found on only one or two different host trees. Fruiting bodies of four species (Coprinopsis strossmayeri, Lentinus tuber-regium, Phallus atrovolvatus, and Psathyrella luteopallida) were found in the forest soil.
Fungal strains were isolated from both marine (39 spp. from 139 strains) and terrestrial environments (27 spp. from 212 strains) (). G. candidum was commonly isolated from marine environments, while T. reesei was more abundant in the terrestrial environment. Eight species, including P. citrinum and T. reesei, were isolated from both environments; however, most species were isolated from either marine (31 spp.) or terrestrial (19 spp.) environments. Various species were found in the seaweed (27 spp. from 85 strains), sea sand (18 spp. from 54 strains), and rotten wood (27 spp. from 212 strains) (). Aspergillus brunneoviolaceus, A. neoniger, G. candidum, P. citrinum, and T. reesei were the species shared across all sources, while many were unique to each source: seaweed (20 spp.), sea sand (10 spp.), and rotten wood (19 spp.). G. candidum, P. citrinum, and T. reesei were the dominant species residing in seaweed, sea sand, and rotten wood, respectively ().
3.3. Species inventory update
By analyzing previous records of fungi in the FSM, dating from 1913, we made a list of 236 species from Yap, Chuuk, Pohnpei, and Kosrae (Table S1). Only 58 of these had been identified using sequence data. The listed fungal species were detected in various sources, including diseased plants (101 spp.), rotting wood (66 spp.), house dust (44 spp.), lichens (14 spp.), and forest soil (3 spp.). Some species (8 spp.) from the marine environment were identified using metagenomics. Most of the listed species belonged to Ascomycota (144 spp., 61.0%) and Basidiomycota (90 spp., 38.1%), while three and one species belonged to Mucoromycota and Glomeromycota, respectively (Table S1).
We added 78 new species to the FSM fungal inventory, resulting in a total of 314 fungal species as follows: Ascomycota (n = 193, 61.5%), Basidiomycota (n = 117, 37.3%), Mucoromycota (n = 3, 1.0%), and Glomeromycota (n = 1, 0.3%).
4. Discussion
We conducted a survey of fungal diversity at Chuuk and Kosrae states in the FSM. Based on molecular and morphological identification, we identified a total of 99 fungal species from various environments. Although 236 fungal species have been previously reported in the FSM, sequence information is available for only a few species [Citation4,Citation16,Citation17]. Morphological features are not enough to identify most fungal species at the species level because of their similar morphological characteristics and variations depending on the environment [Citation17,Citation37–40]. Therefore, we verified the identification of the 99 species collected in this study by performing a thorough sequence analysis, in addition to morphological identification. The morphology-based identifications are currently being verified by sequence-based identification. Sequence analysis plays an important role in distinguishing morphologically similar species and reducing the rate of misidentification. Hence, it is necessary to develop a standardized method for each taxon, based on sequencing, and construct a reliable global database [Citation41]. The sequence information generated in this study will be useful to researchers for the accurate identification of various fungi and for conducting further studies on the fungal diversity of the FSM.
Environmental filtering by hosts, habitats, and sources is an important factor influencing fungal communities [Citation42–44]. In this study, although the sample size was small, many fruiting bodies showed host preference. A previous study has also reported that some fruiting bodies (polypore fungi) in the FSM show host and habitat specificity [Citation7]. The fungal diversity in our study also varied according to the habitat and source. Only eight of the 58 isolated fungal species were commonly found in both marine and terrestrial habitats, while most of the species were unique to their sources: sea sand, seaweed, and wood. We also found that many species isolated from the marine environment were previously not recorded in the FSM. The discovery of many unrecorded species in our study may be due to the inclusion of new environments in the survey. Recent studies have shown that various fungal species inhabit marine environments [Citation45–47]. Annulohypoxylon stygium, an unrecorded species, was isolated only from seaweeds in this study. It produces various secondary metabolites and enzymes [Citation48,Citation49] and provides nutrients to Tremella fuciformis for the growth and development of fruiting bodies [Citation50]. Recently, A. stygium isolated from seaweed has been shown to produce novel molecules that can be used as UV filters [Citation51]. Therefore, investigating various environments may lead to the discovery of industrially and medicinally useful fungal species.
Though the FSM is a small country, it is known as a biodiversity hotspot with high plant and animal diversity. Despite this, there has been relatively little research on its fungal diversity. Although we surveyed only a limited number of sites and sources, we confirmed the presence of 99 fungal species, including at least 78 previously unrecorded species in the FSM, based on morphological features and sequence information. In addition, we constructed a new fungal inventory of the FSM, along with the results of previous studies. Further extensive research across both marine and terrestrial environments should be conducted to discover new fungal species. This study will be an important basis for the discovery of new bioresources and the monitoring of fungi based on ecological changes caused by urban development in the FSM.
Supplemental Material
Download MS Excel (46.2 KB)Acknowledgements
We are grateful to the Chuuk and Kosrae State Government, the Federated States of Micronesia, for allowing marine organism research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- GBIF. GBIF Home Page; 2021. Available from: https://www.gbif.org
- New Zealand Fungarium – Te Kohinga Hekaheka o Aotearoa (PDD) 2021. Specimen data. Accessed through Systematics Collection Data. Available from: http://scd.landcareresearch.co.nz
- Aime MC, Kijpornyongpan T, Abbasi M, et al. A new species of Cintractiella (Ustilaginales) from the volcanic island of Kosrae, Caroline Islands, Micronesia. MC. 2018;42:1–6.
- Cannon P, Klopfenstein NB, Kim M-S, et al. Characterizing forest root‐and butt‐rot fungi in Yap, Palau, Pohnpei, Kosrae, Guam and Saipan. In: Cannon P, editor. Forest pathology in Yap, Palau, Pohnpei, Kosrae, Guam and Saipan, Sep 2013. Vallejo, CA: US Department of Agriculture, Forest Service, Region 5, Forest Health Protection; 2014. p. 38–49.
- Díaz-Valderrama JR, Nguyen HD, Aime MC. Wallemia peruviensis sp. nov., a new xerophilic fungus from an agricultural setting in South America. Extremophiles. 2017;21(6):1017–1025.
- Dixon LJ, Schlub RL, Pernezny K, et al. Host specialization and phylogenetic diversity of Corynespora cassiicola. Phytopathology. 2009;99(9):1015–1027.
- Gilbert GS, Gorospe J, Ryvarden L. Host and habitat preferences of polypore fungi in Micronesian tropical flooded forests. Mycol Res. 2008;112(Pt 6):674–680.
- González-Ball R, Ono Y. Rust fungi (Uredinales) found in Marshall Islands and Pohnpei. Mycoscience. 1998;39(2):221–222.
- Hirooka Y, Tanney JB, Nguyen HD, et al. Xerotolerant fungi in house dust: taxonomy of Spiromastix, Pseudospiromastix and Sigleria gen. nov. in Spiromastigaceae (Onygenales, Eurotiomycetes). Mycologia. 2016;108(1):135–156.
- McKenzie EH, Jackson GV. The fungi, bacteria and pathogenic algae of the Federated States of Micronesia. Technical Papers. South Pacific Commission. 1990.
- Miyazaki Y, Hiraide M, Shibuya H. Molecular cloning of functional genes for high growth-temperature and salt tolerance of the basidiomycete Fomitopsis pinicola isolated in a mangrove Forest in Micronesia. Biosci Biotechnol Biochem. 2007;71(1):273–278.
- Nguyen HD, Jancic S, Meijer M, et al. Application of the phylogenetic species concept to Wallemia sebi from house dust and indoor air revealed by multi-locus genealogical concordance. PLOS One. 2015;10(3):e0120894.
- Redecker D, Raab P, Oehl F, et al. A novel clade of sporocarp-forming species of glomeromycotan fungi in the Diversisporales lineage. Mycol Prog. 2007;6(1):35–44.
- Stewart J, Kim M-S, Ota Y, et al. Phylogenetic and population genetic analyses reveal three distinct lineages of the invasive brown root-rot pathogen, Phellinus noxius, and bioclimatic modeling predicts differences in associated climate niches. Eur J Plant Pathol. 2020;156(3):751–766.
- Tanney JB, Nguyen HD, Pinzari F, et al. A century later: rediscovery, culturing and phylogenetic analysis of Diploöspora rosea, a rare onygenalean hyphomycete. Antonie Van Leeuwenhoek. 2015;108(5):1023–1035.
- Tanney JB, Visagie CM, Yilmaz N, et al. Aspergillus subgenus Polypaecilum from the built environment. Stud Mycol. 2017;88:237–267.
- Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78:63–139.
- Wong M-H, Crous PW, Henderson J, et al. Phyllosticta species associated with freckle disease of banana. Fungal Divers. 2012;56(1):173–187.
- Woudenberg JHC, Meijer M, Houbraken J, et al. Scopulariopsis and scopulariopsis-like species from indoor environments. Stud Mycol. 2017;88:1–35.
- Hawksworth DL. The fungal dimension of biodiversity – magnitude, significance, and conservation. Mycol Res. 1991;95(6):641–655.
- Carlile MJ, Watkinson SC, Gooday GW. The fungi. 2nd ed. San Diego, CA: Elsevier-Academic Press; 2001.
- Gilbertson RL, Ryvarden L. North American polypores. Vol. 1. Abortiporus-Lindtneria. North american polypores. Oslo: Fungiflora A/S; 1986.
- Breitenbach J, Kränzlin F. Fungi of Switzerland. Vol. 1, Ascomycetes. 1984; Lucerne: Verlag Mykologia.
- Breitenbach J, Kränzlin F. Fungi of Switzerland, vol. 2, non-gilled fungi. Lucerne: Verlag Mykologia; 1986.
- Breitenbach J, Kränzlin F. Fungi of Switzerland, vol. 3, agarics 1st part. Lucerne: Verlag Mykologia; 1991.
- Breitenbach J, Kränzlin F. Fungi of Switzerland, vol. 4, agarics 2nd part. Lucerne: Verlag Mykologia; 1995.
- Breitenbach J, Kränzlin F. Fungi of Switzerland, vol. 5, agarics 3rd part. Lucerne: Verlag Mykologia; 2000.
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. Dordrecht: Springer; 1994. p. 183–190.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts . Mol Ecol. 1993;2(2):113–118.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a Guide to Methods and Applications. 1990;18(1):315–322.
- Peterson SW, Vega FE, Posada F, et al. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97(3):659–666.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- O’Donnell K, Kistler HC, Cigelnik E, et al. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci. 1998;95(5):2044–2049.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120.
- Raja HA, Miller AN, Pearce CJ, et al. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod. 2017;80(3):756–770.
- Slepecky RA, Starmer WT. Phenotypic plasticity in fungi: a review with observations on Aureobasidium pullulans. Mycologia. 2009;101(6):823–832.
- Marin-Felix Y, Groenewald J, Cai L, et al. Genera of phytopathogenic fungi: GOPHY 1. Stud Mycol. 2017;86:99–216.
- Hofstetter V, Buyck B, Eyssartier G, et al. The unbearable lightness of sequenced-based identification. Fungal Divers. 2019;96(1):243–284.
- Hibbett D, Abarenkov K, Kõljalg U, et al. Sequence-based classification and identification of fungi. Mycologia. 2016;108(6):1049–1068.
- Baakza A, Vala A, Dave B, et al. A comparative study of siderophore production by fungi from marine and terrestrial habitats. J Exp Mar Biol Ecol. 2004;311(1):1–9.
- Dini-Andreote F, Pylro VS, Baldrian P, et al. Ecological succession reveals potential signatures of marine-terrestrial transition in salt marsh fungal communities. ISME J. 2016;10(8):1984–1997.
- Park MS, Lee JW, Kim SH, et al. Penicillium from rhizosphere soil in terrestrial and coastal environments in South Korea. Mycobiology. 2020;48(6):431–442.
- Godinho VM, de Paula MTR, Silva DAS, et al. Diversity and distribution of hidden cultivable fungi associated with marine animals of Antarctica. Fungal Biol. 2019;123(7):507–516.
- Park MS, Oh S-Y, Fong JJ, et al. The diversity and ecological roles of Penicillium in intertidal zones. Sci Rep. 2019;9(1):1–11.
- Picard KT. Coastal marine habitats harbor novel early-diverging fungal diversity. Fungal Ecol. 2017;25:1–13.
- Cheng M-J, Wu M-D, Chen J-J, et al. Secondary metabolites from the endophytic fungus Annulohypoxylon stygium BCRC 34024. Chem Nat Compd. 2014;50(2):237–241.
- Robl D, dos Santos Costa P, Büchli F, et al. Enhancing of sugar cane bagasse hydrolysis by Annulohypoxylon stygium glycohydrolases. Bioresour Technol. 2015;177:247–254.
- Deng Y, van Peer AF, Lan F-S, et al. Morphological and molecular analysis identifies the associated fungus (“xianghui”) of the medicinal white jelly mushroom, Tremella fuciformis, as Annulohypoxylon stygium. Int J Med Mushrooms. 2016;18(3):253–260.
- Maciel OMC, Tavares RSN, Caluz DRE, et al. Photoprotective potential of metabolites isolated from algae-associated fungi Annulohypoxylon stygium. J Photochem Photobiol B. 2018;178:316–322.