Abstract
The Aspergilli of the section Nidulantes series Versicolores are among the most recurrent molds in indoor environments. These species cause damage to the quality of air. Indeed, they are responsible for allergies, aggravation of asthma and can even cause infections in immunocompromised patients. Molds belonging to the Versicolores series also produce sterigmatocystin, a mycotoxin classified as potential human carcinogen by the International Agency for Research on Cancer (group 2B). Here, we provide for the first time the genome of three species of the series Versicolores: Aspergillus creber, Aspergillus jensenii and Aspergillus protuberus which are the most abundant species of this series in bioaerosols. The genomes of these three species could be assembled with a percentage of completeness of 97.02%, 96.21% and 95.35% for Aspergillus creber, A. jensenii and A. protuberus respectively. These data will allow to study the genes and gene clusters responsible for the expression of virulence factors, the biosynthesis of mycotoxins and the proliferation of these ubiquitous and recurrent molds.
Keywords:
Aspergillus creber (Jurjević, S.W. Peterson & B.W. Horn), A. jensenii (Jurjević, S.W. Peterson & B.W. Horn) and A. protuberus (Munt.-Cvetk) are three airborne Aspergilli belonging to the section Nidulantes series Versicolores [Citation1,Citation2].
Aspergillus creber is considered to be the most abundant mold of the series Versicolores especially in damp indoor environments [Citation3]. It is recurrently found in indoor air [Citation4,Citation5] but it has also been isolated from dust, soil, food (grape, cocoa powder, etc) and animal hair [Citation1,Citation6]. A. jensenii is also commonly found in indoor environment even though it is less recurrent than A. creber [Citation3,Citation5]. It has been isolated from indoor air samples, dust and food (pilled millet) [Citation5,Citation6]. A. protuberus is less recurrent than A. creber and A. jensenii [Citation5] but remains quite frequently found in dust, indoor air and food (brined meat) [Citation1,Citation6]. These three species were also found in clinical samples (arm skin, bronchoalveolar lavage fluid, eye, nail, skin mucosa, sputum and in vaginal discharge) [Citation7–9]. They are considered as cryptic species [Citation10] that can be found in cases of onychomycosis (both A. creber, A. jensenii and A. protuberus) [Citation8] and in endophthalmitis, keratitis, scalp mycosis and in vaginitis (A. protuberus) [Citation9,Citation11,Citation12]. Although these species are opportunistic pathogens, they are also known to be involved in allergies [Citation13], asthma aggravations [Citation14] and may cause infections in immunocompromised patients [Citation15,Citation16]. A. creber, A. jensenii and A. protuberus produce sterigmatocystin [Citation3], a mycotoxin classified as a group 2B (potential human carcinogen) by IARC [Citation17]. This is the first genome report for these three species.
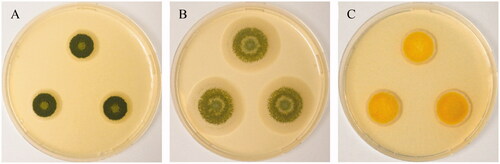
Pure cultures of Aspergillus creber isolate HOSP050413_5_135 and A. protuberus isolate HOSP050413_4_129 were recovered from bioaerosols collected in April 2013 from the fifth and fourth floors of a cancer treatment center (Center François Baclesse, Caen), respectively (). Bioaerosols were collected using a cyclonic biocollector (Bertin Technologies, Montigny-le-Bretonneux, France) during 40 min at 300 L.min−1. Samples were cultured on Malt Extract Agar (MEA) medium supplemented with 0.02% chloramphenicol (Cooper, Melun, France). Plates were incubated at 25 °C and checked daily. Each isolate was purified on the same medium. Pure culture of Aspergillus jensenii isolate C4_18042019 was recovered from the scalp of a patient at Caen University Hospital in April 2019 () for which a mycological examination was prescribed. After inoculation on sabouraud dextrose agar with chloramphenicol and gentamicin (Bio-Rad, Marnes-la-Coquette, France), this isolate was also purified on MEA.
Figure 1. Two weeks-old colonies on Malt Extract Agar, from left to right (A) Aspergillus creber (HOSP050413_5_135), (B) Aspergillus protuberus (HOSP050413_4_129) and (C) Aspergillus jensenii (C4_18042019).
All isolates were molecularly characterized by the primer set Bt2a/b (Bt2a: 5′ GGT AAC CAA ATC GGT GCT GCT TTC 3′ and Bt2b: 5′ ACC CTC AGT GTA GTG ACC CTT GGC 3′ (Eurogentec, Liège, Belgique)). Genomic DNA extracted using the Nucleospin™ Plant II kit (Macherey-Nagel, Duren, Germany) was then sequenced on an Illumina NovaSeq 6000 platform using 2 × 150 bp sequence mode. The raw reads were trimmed using Trimmomatic (version 0.38.0) [Citation18]. Quality-passed reads were assembled using SPAdes pipeline (version 3.12.0) [Citation19] de novo genome assembler with default options. Using BUSCO (Benchmarking Universal Single-Copy Orthologs) pipeline (version 5.2.2) with the Ascomycota odb10 lineage dataset, we estimated the completeness of our genomes [Citation20]. Gene prediction was performed using Augustus (version 3.2.3) [Citation21] using default options. Bandage Info (version 0.8.1) [Citation22] and Quast (version 5.0.2) [Citation23] were used to determine the statistics of de novo assembly graphs and to provide information on the quality of genome assembly, respectively. All data on the assembled genomes are shown in .
Table 1. Summary of the genome assembly and annotation of Aspergillus creber (HOSP050413_5_135), Aspergillus jensenii (20190418_C4) and A. protuberus (HOSP050413_4 129).
These genomic resources will allow comparative genomic analysis to be made among these three recurrent environmental molds and will increase the data available to study the molecular basis of pathogenicity and metabolites production in these organisms. The draft genome sequences of A. creber HOSP050413_5_135, A. jensenii C4_18042019 and A. protuberus HOSP050413_4_129 have been deposited in GenBank under the accession number JAJAEB000000000, JAJAED000000000 and JAJAEC000000000, BioProject number PRJNA768996, PRJNA769000 and PRJNA768998, BioSample number SAMN22074031, SAMN22074134 and SAMN22074035 respectively.
Acknowledgments
ABTE-ToxEMAC is a member of the FHU (University Hospital Federation) RESPIRE: “Pathogènes, ENvironnement et Hôte: une approche intégrative en santé respiratoire”.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jurjevic Z, Peterson SW, Horn BW. Aspergillus section versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 2012;3(1):59–79.
- Houbraken J, Kocsubé S, Visagie CM, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169.
- Jakšić D, Sertić M, Kifer D, et al. Fungi and their secondary metabolites in water-damaged indoors after a major flood event in eastern Croatia. Indoor Air. 2021;31(3):730–744.
- Jakšić Despot D, Šegvić Klarić M. A year-round investigation of indoor airborne fungi in Croatia. Arh Hig Rada Toksikol. 2014;65(2):209–218.
- Géry A, Rioult J-P, Heutte N, et al. First characterization and description of Aspergillus series versicolores in French bioaerosols. JoF. 2021;7(8):676.
- Kobayashi N, Kubosaki A, Takahashi Y, et al. Distribution of sterigmatocystin-producing Aspergilli in Japan. Food Saf (Tokyo). 2018;6(2):67–73.
- Tsang C-C, Hui TWS, Lee K-C, et al. Genetic diversity of Aspergillus species isolated from onychomycosis and Aspergillus hongkongensis sp. nov., with implications to antifungal susceptibility testing. Diagn Microbiol Infect Dis. 2016;84(2):125–134.
- Siqueira JPZ, Sutton DA, García D, et al. Species diversity of Aspergillus section Versicolores in clinical samples and antifungal susceptibility. Fungal Biol. 2016;120(11):1458–1467.
- Borsa BA, Özgün G, Houbraken J, et al. The first case of persistent vaginitis due to Aspergillus protuberus in an immunocompetent patient. Mikrobiyol Bul. 2015;49(1):130–134.
- Vidal-Acuña MR, Ruiz M, Torres MJ, et al. Prevalence and in vitro antifungal susceptibility of cryptic species of the genus Aspergillus isolated in clinical samples. Enferm Infecc Microbiol Clin (Engl Ed). 2019;37(5):296–300.
- Jia J, Chen M, Mo X, et al. The first case report of kerion-type scalp mycosis caused by Aspergillus protuberus. BMC Infect Dis. 2019;19(1):506.
- Al-Hatmi AMS, Castro MA, de Hoog GS, et al. Epidemiology of Aspergillus species causing keratitis in Mexico. Mycoses. 2019;62(2):144–151.
- Rick E, Woolnough K, Pashley C, et al. Allergic fungal airway disease. J Investig Allergol Clin Immunol. 2016;26(6):344–354.
- Tiotiu AI, Novakova P, Nedeva D, et al. Impact of air pollution on asthma outcomes. IJERPH. 2020;17(17):6212.
- Spellberg B, Edwards J, Ibrahim A. Novel perspectives on mucormycosis: pathophysiology, presentation, and management. Clin Microbiol Rev. 2005;18(3):556–569.
- Gletsou E, Ioannou M, Liakopoulos V, et al. Aspergillosis in immunocompromised patients with haematological malignancies. J BUON off J Balk Union Oncol. 2018;23:7–10.
- IARC monographs on the identification of carcinogenic hazards to humans. Available online: https://monographs.iarc.who.int/. (accessed on 10 September 2021).
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120.
- Bankevich A, Nurk S, Antipov D, et al. SPADES: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477.
- Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol Clifton NJ. 2019;1962:227–245.
- Nachtweide S, Stanke M. Multi-genome annotation with AUGUSTUS. Methods Mol Biol Clifton NJ. 2019;1962:139–160.
- Wick RR, Schultz MB, Zobel J, et al. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–3352.
- Mikheenko A, Prjibelski A, Saveliev V, et al. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34(13):i142–50.
