Abstract
Trichoderma fungi have been intensively studied for mycoparasitism, and the latter is closely related to their cell-wall degrading enzymes including chitinase. Here, we studied marine-derived Trichoderma spp., isolated from distinct sources and locations, for chitinolytic and antifungal activity. Based on morphological and phylogenetic analyses, two strains designated GJ-Sp1 and TOP-Co8 (isolated from a marine sponge and a marine alga, respectively) were identified as Trichoderma bissettii. This species has recently been identified as a closely related species to Trichoderma longibrachiatum. The extracellular crude enzymes of GJ-Sp1 and TOP-Co8 showed activities of chitobiosidase and β-N-acetylglucosaminidase (exochitinase) and chitotriosidase (endochitinase). The optimum chitinolytic activity of the crude enzymes was observed at 50 °C, pH 5.0, 0–0.5% NaCl concentrations, and the activities were stable at temperatures ranging from 10 to 40 °C for 2 h. Moreover, the crude enzymes showed inhibitory activity against hyphal growth of two filamentous fungi Aspergillus flavus and Aspergillus niger. To the best of our knowledge, this is the first report of the chitinolytic and antifungal activity of T. bissettii.
1. Introduction
Although some species are pathogenic to cause green mold disease in mushrooms [Citation1], fungi in the genus Trichoderma are versatile plant symbionts and biocontrol agents with beneficial features; these features include mycoparasitic activities and plant growth promotion [Citation2].
In terrestrial ecosystems, Trichoderma spp. are ubiquitous inhabitants of soil and decaying wood [Citation3]. Relative to terrestrial species, studies on marine-derived Trichoderma spp. are limited. Trichoderma isolates from marine environments have gained increasing attention due to their ability to produce diverse bioactive secondary metabolites and enzymes [Citation4,Citation5].
The biocontrol ability of Trichoderma spp. against plant pathogenic fungi is closely related to chitinase production in these spp. [Citation6,Citation7]. Chitinases are present in bacteria, fungi, insects, plants, and vertebrates including humans, and vary in their chemical structure, substrate specificity, and mode of action [Citation8]. In addition to their biological functions in nutrition, fungal morphogenesis, insect growth, plant development, and host defense against chitin-containing pathogens [Citation9,Citation10], chitinases have several potential applications. For example, because fungal cell walls and insect exoskeletons consist of chitin, chitinases can be utilized to control phytopathogenic fungi and insects [Citation11]. In addition, chitinase can be used for the treatment of chitinous waste from the sea food industry. Moreover, some chitinases and chitin oligomers have antimicrobial, immunopotentiating, and anticancer activities [Citation12–15].
Several Trichoderma species including Trichoderma asperellum, Trichoderma atroviride, Trichoderma hazianum, Trichoderma koningiopsis, Trichoderma longibrachiatum, Trichoderma virens, and Trichoderma viride are known to produce chitinases [Citation16–22]. In marine-derived Trichoderma species, algae-derived Trichoderma lixii IG127, mangrove soil-derived T. viride AUMC 13021, and marine sponge-derived Trichoderma asperelloides O.Y. 1607 and T. atroviride O.Y. 3807 are known to possess chitinolytic activities [Citation23–25]. Notably, chitinases purified from the T. viride AUMC 13021 strain exhibit anticancer activity [Citation23].
In this paper, we studied chitinolytic and antifungal activities of two marine-derived Trichoderma bissettii strains. Using the extracellular crude enzymes of GJ-Sp1 and TOP-Co8, we investigated production of endo- and exo-chitinases, the optimum conditions and thermal stability for chitinolytic activity, and inhibitory effect on fungal hyphal growth.
2. Materials and methods
2.1. Fungal isolation and culture conditions
The marine sponge Hymeniacidon sinapium and blades of a marine alga Codium sp. were collected from Gijang-gun, Busan, Republic of Korea (35.11′30.22″N, 129.13′29.75″E) and Muan-gun, Jeollanam-do, Republic of Korea (34.58′05.5″N, 126.23′10.3″E), respectively. The marine sponge and the marine alga Codium sp. were collected from rock surfaces in a tidal pool (at ∼15 cm depth) and seashore sand, respectively. We stored the marine samples in plastic bags, carried to the laboratory, and used for isolation of fungi within 12 h. These marine samples were washed with sterile distilled water and cut into ∼1 cm long pieces using a surgical blade. The segments were placed on potato dextrose agar (PDA; BD Difco, Sparks, MD, USA) and yeast mold agar (YM, BD) containing ampicillin (0.01%, w/v) (Sigma-Aldrich, St. Louis, MO, USA), streptomycin (0.01%, w/v) (Sigma-Aldrich), and NaCl (3%, w/v), and incubated at 20 °C for 7–14 d. Individual fungal colonies were isolated and transferred to fresh media. After isolation, fungal strains were cultured on PDA at 25 °C unless otherwise described. The strains isolated from marine sponges and marine algae were designated as GJ-Sp1 and TOP-Co8, respectively.
2.2. Microscopy
GJ-Sp1 and TOP-Co8 strains were cultured on agar plugs of Czapek-Dox agar (BD) at 25 °C for 5 d [Citation26]. The morphology of spores and conidiophores was observed using a Leica CTR6000 microscope equipped with a Leica DMC2900 camera (Leica, Wetzlar, Germany). Image acquisition and processing were conducted using LASV4.5 software (Leica).
2.3. DNA extraction, sequencing, and phylogenetic analysis
To extract DNA, spores of GJ-Sp1 and TOP-Co8 were inoculated in PDB and cultured at 25 °C, 200 rpm for 5 days. Mycelia were harvested and genomic DNA (gDNA) was isolated using a 5 min mushroom DNA extraction kit (BioFactories, Daejeon, Korea). Polymerase chain reaction (PCR) was conducted using the primers ITS1 and ITS4 to amplify the nuclear ribosomal DNA internal transcribed spacer (ITS) [Citation27], using the primers EF1-728F [Citation28], and TEF1LLErev [Citation29] to amplify the tef1α gene encoding a translation elongation factor 1α. PCR conditions were as follows: 3 min at 95 °C; 34 cycles of 15 s at 95 °C, 30 s at 58 °C, and 1 min at 72 °C, and finally post-polymerization at 72 °C for 15 min. PCR products were purified using a Gel Extraction Kit (Qiagen, Hilden, Germany). Sequences of all the PCR products were analyzed by Macrogen (Macrogen, Seoul, Korea).
Closely related sequence matches to the ITS and tef1α sequences of GJ-Sp1 and TOP-Co8 were investigated by BLASTN search in GenBank and based on previously published studies [Citation30]. MEGA version 6 [Citation31] was used to align individual sequences and generate phylogenetic trees using the neighbor-joining method, followed by bootstrapping with 1,000 replicates. The Hypocrea pachybasioides Tr 46 ITS (GenBank DQ112552) and tef1α (GenBank AY750885) sequence were included as an outgroup for phylogenetic analysis [Citation30].
2.4. Plate assay for chitinolytic activity
Spores were collected from each isolate cultured on PDA at 25 °C for 7–14 d. Then, 10 µL of spore suspension was inoculated on the center of media containing 2% (w/v) colloidal chitin, 0.1% (w/v) (NH4)2SO4, 0.2% (w/v) KH2PO4, 0.3% (w/v) Na2HPO4, 0.05% (w/v) MgSO4·7H2O, 0.02% (w/v) yeast extract. Colloidal chitin was made as previously described [Citation32]. Following 4–14 d of incubation at 25 °C, chitinolytic activity was assessed based on the presence of clear zones around colonies.
2.5. Preparation of extracellular crude enzyme
Marine-derived fungal strains were cultured on PDA at 25 °C for 7 d. Ten agar plugs from the PDA plates were removed using a cork borer (10 mm in diameter) and inoculated into liquid basal media [Citation32] containing 1% (w/v) colloidal chitin. Following a 14 d incubation at 25 °C, the supernatant was separated from fungal tissue using a sterile Miracloth (Merck, New Jersey, USA) and sterilized using syringe filters with a 0.2 μm pore size membrane (Corning, Corning, NY, USA). Extracellular crude enzymes were concentrated using Amicon Ultra 3000 nominal molecular weight limit filters (Merck) and were buffer-exchanged with 20 mM sodium phosphate buffer (pH 6.0). Protein quantification in the supernatant was conducted using a Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA, USA).
2.6. Endo- and exo-chitinase activity
To measure the endo- and exo-chitinase activity of crude enzymes of GJ-Sp1 and TOP-Co8, the Chitinase Assay Kit was used (Sigma-Aldrich). The experimental procedures were conducted according to the manufacturer’s instructions. A crude enzyme solution containing 0.5 μg protein was used to hydrolyze three individual substrates (4-nitrophenyl N,N′-diacetyl-β-D-chitobioside, 4-nitrophenyl N-acetyl-β-D-glucosaminide, and 4-nitrophenyl β-D-N,N′N″-triacetylchitotriose). When each substrate is hydrolyzed, p-nitrophenol is released and detected by ionization in basic pH at 405 nm. One unit was defined as the amount of enzyme required to release 1.0 μmol of p-nitrophenol from the appropriate substrate per minute at pH 4.8, at 37 °C. This experiment was performed in three replicates.
2.7. 3,5-Dinitrosalicylic acid (DNS) assay
Distinct from the kit assay using specific substrates containing a 4-nitrophnol group, colloidal chitins were used as substrates for chitinolytic activity in a DNS assay. The substrate solutions could be prepared at specific pHs and NaCl concentrations more easily at the DNS assay compared to the kit assay due to the distinct substrates. Chitinolytic activity of GJ-Sp1 and TOP-Co8 was measured using a DNS assay [Citation33] with 2% (w/v) colloidal chitin as the substrate. A crude enzyme solution containing 30 μg of protein was used to examine the chitinolytic activity by a DNS assay. A standard curve was obtained using N-acetylglucosamine (GlcNAc, Sigma) dissolved in 20 mM phosphate buffer (pH 6.0). The optical densities of reaction mixtures were measured at 545 nm against the substrate and enzyme blanks [Citation32]. Chitinolytic activity was defined as the amount of GlcNAc (μg/mL) produced by 1 mg of protein in the crude enzyme per hour under tested conditions. All experiments to evaluate chitinolytic activity of GJ-Sp1 and TOP-Co8 were performed in biological triplicates.
2.8. Optimization of temperature, pH, NaCl concentration, and incubation time for chitinolytic activity
The effect of temperature on chitinolytic activity was examined at 10, 20, 30, 40, 50, and 60 °C. The extracellular crude enzyme was incubated with 1 mL of 2% (w/v) colloidal chitin suspended in 20 mM phosphate buffer (pH 6.0) at each temperature for 2 h. One milliliter of the reaction mixture was used to evaluate chitinolytic activity using the DNS assay.
The effect of pH on chitinolytic activity was studied using 2% (w/v) colloidal chitin solution suspended in buffers at various pH levels (20 mM sodium acetate buffer for pH 4.0 and 5.0; 20 mM sodium phosphate buffer for pH 6.5 and 8.0; 20 mM glycine-NaOH buffer for pH 9.5 and 11). Then, the reaction mixture was incubated at 37 °C for 2 h.
The effect of NaCl concentration on chitinolytic activity was evaluated at 0, 0.5, 1.5, 3.0, and 5.0% (w/v) NaCl. The crude enzyme was incubated with 2% (w/v) colloidal chitin suspended in 20 mM phosphate buffer (pH 6.0) at each NaCl concentration at 37 °C for 2 h. Following the addition of NaCl to the buffer, the pH was adjusted to 6.0.
To study the effect of incubation time on chitinolytic activity, the crude enzyme was incubated with 2% (w/v) colloidal chitin suspended in 20 mM phosphate buffer (pH 6.0) at 37 °C for 15, 30, 60, 120, and 240 min. At each time point, the enzyme reaction was stopped by the addition of 1 mL DNS reagent.
2.9. Thermal stability of chitinolytic activity
The thermal stability of GJ-Sp1 and TOP-Co8 chitinases was examined as previously described with a few modifications [Citation34]. Briefly, a 100 μL solution of the extracellular crude enzyme in 20 mM sodium phosphate buffer was pre-incubated at 10, 25, 40, 50, and 60 °C for 1 or 2 h, and chilled immediately on ice. The crude enzyme was incubated with 1 mL of 2% colloidal chitin solution in 20 mM sodium phosphate buffer (pH 6.0) at 37 °C for 2 h.
2.10. Antifungal activity assay
A microplate-based growth assay was conducted to determine the antifungal activity of extracellular crude enzymes of GJ-Sp1 and TOP-Co8. The target fungal species were Aspergillus flavus (KCCM 60330/ATCC 22546) and Aspergillus niger (KCCM 60332/ATCC 16888) purchased from the Korean Culture Center of Microorganisms (KCCM). The antifungal assay procedure was conducted as previously described with a few modifications [Citation35]. Briefly, 50 μL conidia suspension (2.5 × 104 conidia/mL) in 10% potato dextrose broth (PDB) containing 0.02% (w/v) chloramphenicol was loaded into each well of a 96 well plate. Thereafter, 50 μL crude enzyme containing 10 μg protein was added, resulting in a total reaction volume of 100 μL in each well. For a positive control for fungal growth, 50 μL of 10% PDB containing 0.02% chloramphenicol, instead of the crude enzyme, was added to the fungal culture. For the negative control, no fungal spores were added to each well (crude enzyme only). Following incubation at 25 °C for 48 h, fungal growth was determined using unaided eyes, microscopic observation, and optical density measurement at 600 nm (HIDEX, Finland). This experiment was performed in three replicates.
2.11. Statistical analysis
Data were analyzed using Prism software (version 5.0) (GraphPad Software, San Diego, CA, USA). All experiments were conducted in biological triplicates, and one-way or two-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison tests was performed unless otherwise indicated.
2.12. Accession numbers
The sequence information of the ITS and tef1α of GJ-Sp1 and TOP-Co8 was deposited in GenBank (accession numbers MW295455 and MW295457 for ITS; MW251413 and MW251414 for tef1α).
3. Results
3.1. Identification of marine-derived Trichoderma strains
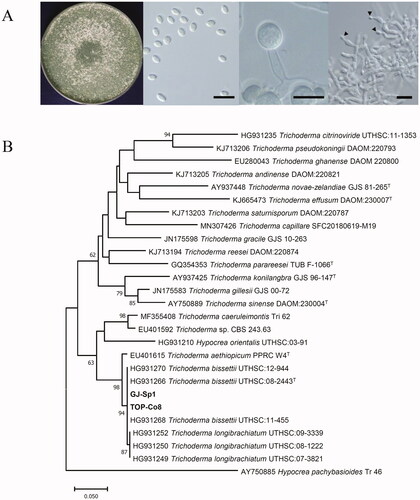
Two marine-derived strains GJ-Sp1 and TOP-Co8 were isolated from distinct sources (marine sponge and marine algae, respectively) collected at distinct locations (Busan and Muan, Jeollanam-do). Both grew readily to form whitish and green granular colonies on PDA at 25 °C for 7 d and produced ellipsoidal conidia with sizes in the range of 2.8–3.3 μm in width × 4.4–5.6 μm in length (n = 10) (). GJ-Sp1 and TOP-Co8 also produced subglobose chlamydospores with sizes in the range of 6.4–8.2 μm in width × 8.2–9.1 μm in length (n = 10) (). Overall, the morphological features of GJ-Sp1 and TOP-Co8 were identical. These strains were selected for further study because their colony, conidiophore, and spore morphology suggested that they were putative Trichoderma species [Citation31,Citation36].
Figure 1. Identification of marine-derived Trichoderma strains. As both strains showed identical morphologies, colony and microscopic images of GJ-Sp1 are presented here to represent both strains. (A) The plate picture shows GJ-Sp1 cultured on potato dextrose agar (PDA) at 25 °C for 7 d. Microscopic images of GJ-Sp1. Conidia (left), chlamydospores (middle), and conidiophores (right) were observed. Black triangles indicate conidia developed from phialides. Scale bar = 10 μm. (B) Phylogenetic analysis of GJ-Sp1 and TOP-Co8. Partial segments of the translation elongation factor 1 α gene (tef1α) were used to generate a neighbor-joining phylogenetic tree. Numbers at nodes indicate percent bootstrap values from 1000 replications (values <50% are not shown). The scale bar indicates the number of nucleotide substitutions per site, and “T” next to the strain name depicts a type specimen of the fungal species.
3.2. Molecular identification of GJ-Sp1 and TOP-Co8
In addition to the morphological analysis for identification of GJ-Sp1 and TOP-Co8, molecular identification was conducted using sequences of ITS and a tef1α gene. The ITS and a tef1α gene sequences of GJ-Sp1 showed 100% identity to those of TOP-Co8. From the BLASTN search, ITS of GJ-Sp1 and TOP-Co8 showed a high degree of similarity to T. bissettii UTHS:09-2160 (100%) and T. longibrachiatum S328 (99.81%). A neighbor-joining phylogenetic tree generated by ITS sequences was not completely resolved to identify GJ-Sp1 and TOP-Co8 (Supplementary Figure 1). Additionally, the BLASTN search revealed that tef1a of GJ-Sp1 and TOP-Co8 showed 100% and 95.11% sequence identity to that of the T. bissettii-type specimen UTHSC:08-2443 (GenBank HG931266) and several T. longibrachiatum strains, respectively. Neighbor-joining phylogenetic analysis using tef1a sequences showed that GJ-Sp1 and TOP-Co8 were grouped with T. bissettii strains (94% degree of confidence) (). Consequently, based on morphological and phylogenetic analyses, both GJ-Sp1 and TOP-Co8 were identified as T. bissettii [Citation31].
3.3. Chitinolytic activity of GJ-Sp1 and TOP-Co8 on colloidal chitin agar
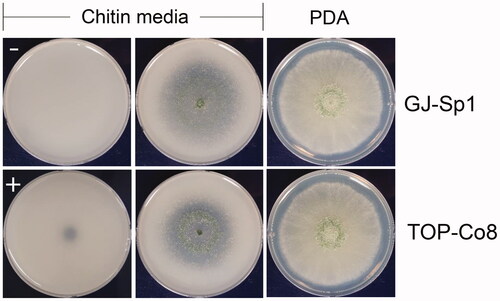
To examine chitinolytic activity of GJ-Sp1 and TOP-Co8, colloidal chitin was used as a substrate. Inoculation of chitinase from T. viride (positive control) changed opaque areas of chitin media to transparent ones (). Inoculation of sterile distilled water (negative control) did not alter the transparency of the media. Thus, we selected chitinolytic fungal strains that showed a transparent peripheral zone of the colony.
Figure 2. Chitinolytic marine-derived fungi GJ-Sp1 and TOP-Co8. Conidia of fungal isolates were inoculated on PDA or colloidal chitin agar, and incubated at 25 °C for 7 d (PDA) or 4 d (chitin media). “−” and “+” indicate negative and positive controls, respectively. Chitinolytic activity was determined by the presence of opaque areas on plates.
3.4. Endo- and exo-chitinase activities of GJ-Sp1 and TOP-Co8
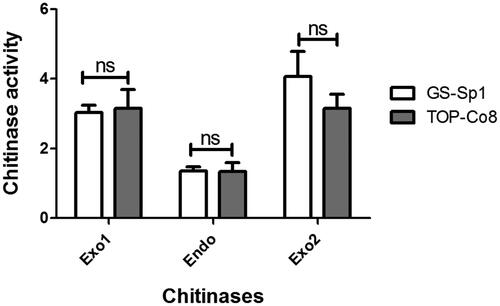
Endo- and exo-chitinase activities of the crude enzyme were assessed using the Chitinase Assay Kit (). The activities of two chitinolytic enzymes (chitobiosidase and β-N-acetylglucosaminidase) were measured for exochitinase activity, and chitotriosidase activity was evaluated for endochitinase activity [Citation37]. Overall, endo- and exo-chitinase activities of GJ-Sp1 were not different from those of TOP-Co8 under all tested conditions. In addition, chitotriosidase (endochitinase) activity showed the lowest levels among the three chitinases in both strains.
Figure 3. Endo- and exo-chitinase activity of the crude enzyme. Using a chitinase assay kit, endo- and exo-chitinase activities were investigated. Exo1 = chitobiosidase (exochitinase); Endo = chitotriosidase (endochitinase); Exo2 = β-N-acetylglucosaminidase (exochitinase). Chitinolytic activity is presented as the mean ± standard deviation of three biological replicates. “ns” indicates statistically “no difference (p > 0.05) between two compared samples” (connected with a horizontal line above the bar graph).
3.5. Optimum temperature, pH, and NaCl concentration for chitinolytic activity
To establish optimum conditions for chitinolytic activity of GJ-Sp1 and TOP-Co8, the extracellular crude enzyme was incubated at various temperatures ranging from 10 to 60 °C (). In both strains, activities increased gradually with increasing temperature from 10 to 50 °C, peaked at 50 °C, and decreased at 60 °C to levels similar to those at 30 °C (p > 0.05 between 30 and 60 °C). The chitinolytic activities of the GJ-Sp1 crude enzyme were not significantly different from those of TOP-Co8 at all examined temperatures (p > 0.05), except at 40 °C. At 40 °C, the activity of the GJ-Sp1 crude enzyme was marginally higher than that of TOP-Co8 (p < 0.05).
Figure 4. Effect of temperature, pH, NaCl concentration, and incubation time on chitinolytic activity of the extracellular crude enzyme. Chitinolytic activity was evaluated using the crude enzyme of GJ-Sp1 and TOP-Co8 using the 3,5-dinitrosalicylic acid (DNS) assay. The crude enzyme was incubated with 2% colloidal chitin at a specific (A) temperature, (B) pH, (C) NaCl concentration, and (D) incubation time. Chitinolytic activity is presented as the mean ± standard deviation of three biological replicates. “ns” and “*” indicate statistically “no difference between samples (p > 0.05)” and “difference between samples (p < 0.05),” respectively.
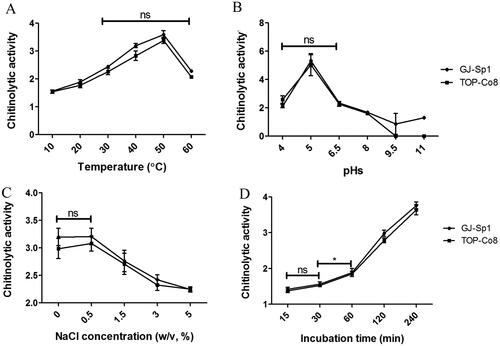
The effect of pH on chitinolytic activity was evaluated at pH 4.0, 5.0, 6.5, 8.0, 9.5, and 11 (). The maximum activity was found at pH 5.0, which was ∼2-fold higher than that at pH 4.0 and 6.5. Chitinolytic activities decreased gradually with increasing pH levels from 6.5 to 9.5. At pH 9.5 and 11, enzyme activities were marginal and not detectable in GJ-Sp1 and TOP-Co8, respectively.
The effect of NaCl concentration on chitinolytic activity was examined at 0, 0.5, 1.5, 3.0, and 5.0% NaCl (w/v) (). The maximum activity observed at 0 and 0.5% NaCl was not significantly different (p > 0.05), and decreased thereafter. There was no difference in chitinolytic activity under all tested NaCl concentrations between GJ-Sp1 and TOP-Co8 (p > 0.05).
The effect of incubation time on chitinolytic activity was investigated at 15, 30, 60, 120, and 240 min (). The activities were not significantly different between 15 and 30 min (p > 0.05) and increased with prolonged incubation time in both strains. There was no difference in chitinolytic activity under all tested incubation times between GJ-Sp1 and TOP-Co8, except for 120 min, which exhibited marginally higher activity in GJ-Sp1 than in TOP-Co8.
3.6. Chitinolytic activity of GJ-Sp1 and TOP-Co8 at optimum conditions
Based on the aforementioned single factor experimental results, the optimum conditions for chitinolytic activities of crude enzymes of GJ-Sp1 and TOP-Co8 were 50 °C, pH 5.0, 0–0.5% NaCl, and 240 min incubation. Under combined optimum conditions, chitinolytic activities were examined using 1, 2, and 4% (w/v) colloidal chitin as substrates. The activities increased gradually with increasing concentrations of colloidal chitin from 1 to 4% (). Chitinolytic activities at combined optimum conditions appeared higher than the activities from any single factor experiments by 1.3–2.2-fold (GJ-Sp1) and 1.2–1.9-fold (TOP-Co8) (). There was no significant difference in chitinolytic activity between GJ-Sp1 and TOP-Co8 at 1 and 2% colloidal chitin concentrations (p > 0.05). However, the activity of GJ-Sp1 was higher than that of TOP-Co8 at 4% colloidal chitin concentration (p < 0.05).
Table 1. Chitinolytic activities at combined optimum conditions.
3.7. Thermal stability of the crude enzymes of GJ-Sp1 and TOP-Co8
The thermal stability of crude enzymes was investigated at 10, 25, 40, 50, and 60 °C for 1 or 2 h (). Overall, there was no significant difference between GJ-Sp1 and TOP-Co8 at any of the tested temperatures and pre-incubation times (p > 0.05). When pre-incubated at 10, 25, and 40 °C, the crude enzyme retained 93–99% chitinolytic activity relative to the control (without pre-incubation), showing statistically similar activities at these temperatures (p > 0.05). The activity significantly declined by pre-incubation at 50 °C for 1 or 2 h, accounting for ∼50–70% of the control. Pre-incubation at 60 °C for 1 or 2 h resulted in the decreased chitinolytic activity of the crude enzymes, exhibiting ∼40–44% residual activity.
Table 2. Thermal stability of the extracellular crude enzyme of GJ-Sp1 and TOP-Co8.
3.8. Antifungal activity of the crude enzymes of GJ-Sp1 and TOP-Co8
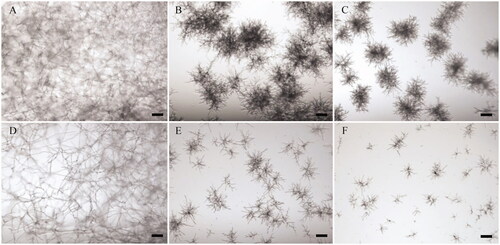
Antifungal activity was assessed against two fungal species, A. flavus and A. niger (target fungi). Following incubation with the crude enzymes for 48 h, growth of target fungi was observed. Both A. flavus and A. niger grew readily and formed mycelia in the control (no addition of the crude enzyme) (). In contrast, the crude enzymes of GJ-Sp1 and TOP-Co8 inhibited hyphal growth of both target fungi compared to that of the control ().
Figure 5. Antifungal activity of the crude enzyme. Conidia of Aspergillus flavus and Aspergillus niger were cultured with the crude enzyme in a 96-well microplate. Following incubation at 25 °C for 2 d, fungal growth was observed using a microscope. (A–C) A. flavus; (D–F) A. niger. (A,D) Positive control for fungal growth (no addition of the crude enzyme); (B,E) Inhibition of hyphal growth by addition of the crude enzymes of GJ-Sp1; (C,F) Inhibition of hyphal growth by addition of the crude enzymes of TOP-Co8. Scale bar = 100 μm.
4. Discussion
In this study, we characterized the chitinolytic activity of two marine-derived T. bissettii strains, GJ-Sp1 and TOP-Co8. T. bissettii was first identified from clinical specimens as a closely related species to T. longibrachiatum [Citation31] and has not been actively studied yet. Recently, this species was isolated from mudflats and showed notable antioxidant activity among other marine-derived Trichoderma spp. [Citation38].
Environmental isolates of fungi often display distinct bioactivities even if they belong to the same species. For example, several isolates of marine-derived Corollospora angusta show different antifungal and cellulolytic enzyme activities [Citation39]. Moreover, four marine-derived Trichoderma guizhouense isolates from fish eggs, a marine alga, mudflat, and sea sand also show different patterns in radical-scavenging, antifungal, and tyrosinase inhibition activities [Citation38]. GJ-Sp1 and Top-Co8 were obtained from distinct isolation sources and locations. However, overall the extracellular crude enzyme of these strains exhibited similar chitinolytic activities under all tested conditions ( and ). Moreover, the crude enzymes of both strains appeared to have an inhibitory effect on the hyphal growth of A. flavus and A. niger (). The inhibitory effect on the growth of A. flavus and A. niger by Trichoderma spp. has been also reported in T. asperellum, T. atroviride, Trichoderma citrinoviride, and Trichoderma harzianum [Citation40–42].
Although experimental designs to optimize chitinase activity vary across previous studies, the optimum conditions of many fungal chitinases are 23–50 °C and pH 4.0–7.6 [Citation32,Citation43–47]. In particular, chitinases of Trichoderma spp., including T. harzianum, T. longibrachiatum, and T. viride, show maximum activities at 50–55 °C and pH 3.5–4.5 [Citation20,Citation22,Citation48], except for T. asperellum chitinases with optimum conditions of 30–40 °C and pH 6.0–7.0 [Citation21,Citation49]. The optimum temperature of the crude enzyme of GJ-Sp1 and TOP-Co8 was 50 °C (), which is similar to that of T. harzianum, T. longibrachiatum, and T. viride. In contrast, the optimum pH of GJ-Sp1 and TOP-Co8 (pH 5.0) was slightly higher than that of the three Trichoderma spp. The optimum NaCl concentration for chitinolytic activity of GJ-Sp1 and TOP-Co8 was 0–0.5%, which is consistent with that of T. harzianum [Citation48] but different from T. asperellum (optimized at 3% NaCl) [Citation49]. Furthermore, the thermal stability of T. bissettii chitinases determined in this study was similar to that of other Trichoderma spp. [Citation20–22,Citation48].
Generally, chitinases are divided into two main groups: endochitinases and exochitinases [Citation50]. The former randomly cleaves chitin at internal sites, resulting in the production of GlcNAc multimers with low molecular mass such as chitotriose, chitobiose, and diacetylchitobiose. The latter produces monomeric or dimeric units of GlcNAc from the non-reducing end, and chitobiosidases and N-acetylglucosaminidases are included in this category [Citation8].
In our data, the crude enzymes of GJ-Sp1 and TOP-Co8 showed endochitinase (chitotriosidase) and exo-chitinase (chitobiosidase and β-N-acetylglucosaminidase) activities (). Trichoderma possesses a variety of chitinolytic enzymes, and the presence of both endo- and exo-chitinases is important for maximum efficacy in cell-wall degrading activity [Citation6]. In addition, each chitinolytic enzyme in Trichoderma spp. produce distinct chitin oligomers with bioactivities. For example, T. harzianum Chit42 chitinase hydrolyzes (GlcNAc)6 into three (GlcNAc)2 units. In contrast, degradation of (GlcNAc)6 by Chit33 chitinase results in the production of (GlcNAc)2 and (GlcNAc)4 [Citation51]. It is noteworthy that we do not have information on how much endo- and exo-chitinases are in the crude enzyme of this study. There is a possibility that the activities of exochitinases were higher than endochitinase () because the crude enzyme contained a higher amount of exochitinases than endochitinase. Therefore, future work would require purification and characterization of individual T. bissettii chitinases for biotechnological applications.
Trichoderma spp. have been extensively studied as biocontrol agents against fungal phytopathogens [Citation52]. Chitinolytic activity of Trichoderma fungi is one of the major determinants of their mycoparasitic ability [Citation6,Citation7]. To the best of our knowledge, this is the first report of chitinolytic activity of T. bissettii, in particular, of marine-derived strains. Our results suggest that T. bissettii has the potential to be utilized as an additional Trichoderma species to produce fungal chitinases and serve as biocontrol agents.
Supplemental Material
Download TIFF Image (316.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Park MS, Bae KS, Yu SH. Two new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology. 2006;34(3):111–113.
- Guzman-Guzman P, Porras-Troncoso MD, Olmedo-Monfil V, et al. Trichoderma species: versatile plant symbionts. Phytopathology. 2019;109(1):6–16.
- Druzhinina IS, Kopchinskiy AG, Kubicek CP. The first 100 Trichoderma species characterized by molecular data. Mycoscience. 2006;47(2):55–64.
- Nicoletti R, Vinale F. Bioactive compounds from marine-derived Aspergillus, Penicillium, Talaromyces and Trichoderma species. Mar Drugs. 2018;16(11):408.
- Su D, Ding L, He S. Marine-derived Trichoderma species as a promising source of bioactive secondary metabolites. Mini Rev Med Chem. 2018;18(20):1702–1713.
- Saba H, Vibhash D, Manisha M, et al. Trichoderma – a promising plant growth stimulator and biocontrol agent. Mycosphere. 2012;3(4):524–531.
- Kubicek CP, Mach RL, Peterbauer CK, et al. Trichoderma: from genes to biocontrol. J Plant Pathol. 2001;83:11–23.
- Rathore AS, Gupta RD. Chitinases from bacteria to human: properties, applications, and future perspectives. Enzyme Res. 2015;2015:791907.
- Hamid R, Khan MA, Ahmad M, et al. Chitinases: an update. J Pharm Bioallied Sci. 2013;5(1):21–29.
- Gooday GW, Zhu WY, O'Donnell RW. What are the roles of chitinases in the growing fungus? FEMS Microbiol Lett. 1992;100(1–3):387–391.
- Okongo RN, Puri AK, Wang Z, et al. Comparative biocontrol ability of chitinases from bacteria and recombinant chitinases from the thermophilic fungus Thermomyces lanuginosus. J Biosci Bioeng. 2019;127(6):663–671.
- Benhabiles M, Salah R, Lounici H, et al. Antibacterial activity of chitin, chitosan, and its oligomers prepared from shrimp shell waste. Food Hydrocoll. 2012;29(1):48–56.
- Pan XQ, Shih CC, Harday J. Chitinase induces lysis of MCF-7 cells in culture and of human breast cancer xenograft B11-2 in SCID mice. Anticancer Res. 2005;25(5):3167–3172.
- Tokoro A, Kobayashi M, Tatewaki N, et al. Protective effect of N-acetyl chitohexaose on Listeria monocytogenes infection in mice. Microbiol Immunol. 1989;33(4):357–367.
- Tsukada K, Matsumoto T, Aizawa K, et al. Antimetastatic and growth-inhibitory effects of N-acetylchitohexaose in mice bearing Lewis lung carcinoma. Jpn J Cancer Res. 1990;81(3):259–265.
- Baek JM, Howell DR, Kenerley CM. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr Genet. 1999;35(1):41–50.
- Baldoni DB, Antoniolli ZI, Mazutti MA, et al. Chitinase production by Trichoderma koningiopsis UFSMQ40 using solid state fermentation. Braz J Microbiol. 2020;51(4):1897–1908.
- de la Cruz J, Hidalgo-Gallego A, Lora JM, et al. Isolation and characterization of three chitinases from Trichoderma harzianum. Eur J Biochem. 1992;206(3):859–867.
- Klemsdal SS, Clarke JL, Hoell IA, et al. Molecular cloning, characterization, and expression studies of a novel chitinase gene (ech30) from the mycoparasite Trichoderma atroviride strain P1. FEMS Microbiol Lett. 2006;256(2):282–289.
- Kovacs K, Szakacs G, Pusztahelyi T, et al. Production of chitinolytic enzymes with Trichoderma longibrachiatum IMI 92027 in solid substrate fermentation. Appl Biochem Biotechnol. 2004;118(1–3):189–204.
- Loc NH, Huy ND, Quang HT, et al. Characterisation and antifungal activity of extracellular chitinase from a biocontrol fungus, Trichoderma asperellum PQ34. Mycology. 2020;11(1):38–48.
- Omumasaba CA, Yoshida N, Ogawa K. Purification and characterization of a chitinase from Trichoderma viride. J Gen Appl Microbiol. 2001;47(2):53–61.
- Abu-Tahon MA, Isaac GS. Anticancer and antifungal efficiencies of purified chitinase produced from Trichoderma viride under submerged fermentation. J Gen Appl Microbiol. 2020;66(1):32–40.
- Gal-Hemed I, Atanasova L, Komon-Zelazowska M, et al. Marine isolates of Trichoderma spp. as potential halotolerant agents of biological control for arid-zone agriculture. Appl Environ Microbiol. 2011;77(15):5100–5109.
- Pasqualetti M, Barghini P, Giovannini V, et al. High production of chitinolytic activity in halophilic conditions by a new marine strain of Clonostachys rosea. Molecules. 2019;24(10):1880.
- Riddell RW. Permanent stained mycological preparations obtained by slide culture. Mycologia. 1950;42(2):265–270.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556.
- Jaklitsch WM, Komon M, Kubicek CP, et al. Hypocrea voglmayrii sp. nov. from the Austrian alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97(6):1365–1378.
- Sandoval-Denis M, Sutton DA, Cano-Lira JF, et al. Phylogeny of the clinically relevant species of the emerging fungus Trichoderma and their antifungal susceptibilities. J Clin Microbiol. 2014;52(6):2112–2125.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739.
- Chung D, Baek K, Bae SS, et al. Identification and characterization of a marine-derived chitinolytic fungus, Acremonium sp. YS2-2. J Microbiol. 2019;57(5):372–380.
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426–428.
- Kwon YM, Choi HS, Lim JY, et al. Characterization of amylolytic activity by a marine-derived yeast Sporidiobolus pararoseus PH-Gra1. Mycobiology. 2020;48(3):195–203.
- Garrigues S, et al. Three antifungal proteins prom Penicillium expansum: different patterns of production and antifungal activity. Front Microbiol. 2018;9:2370.
- Jaklitsch WM, Voglmayr H. Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and macaronesia. Stud Mycol. 2015;80:1–87.
- Elmonem MA, van den Heuvel LP, Levtchenko EN. Immunomodulatory effects of chitotriosidase enzyme. Enzyme Res. 2016;2016:2682680.
- Kim K, Heo YM, Jang S, et al. Diversity of Trichoderma spp. in marine environments and their biological potential for sustainable industrial applications. Sustainability. 2020;12(10):4327.
- Hong J-H, Jang S, Heo YM, et al. Investigation of marine-derived fungal diversity and their exploitable biological activities. Mar Drugs. 2015;13(7):4137–4155.
- Stracquadanio C, Quiles JM, Meca G, et al. Antifungal activity of bioactive metabolites produced by Trichoderma asperellum and Trichoderma atroviride in liquid medium. JOF. 2020;6(4):263.
- Sadykova VS, Kurakov AV, Kuvarina AE, et al. Antimicrobial activity of fungi strains of Trichoderma from Middle siberia. Appl Biochem Microbiol. 2015;51(3):355–361.
- Leelavathi MS, Vani L, Reena P. Antimicrobial activity of Trichoderma harzianum against bacteria and fungi. Int J Curr Microbiol App Sci. 2014;3:96–103.
- Guo RF, Shi BS, Li DC, et al. Purification and characterization of a novel thermostable chitinase from Thermomyces lanuginosus SY2 and cloning of its encoding gene. Agric Sci China. 2008;7(12):1458–1465.
- van Munster JM, van der Kaaij RM, Dijkhuizen L, et al. Biochemical characterization of Aspergillus niger CfcI, a glycoside hydrolase family 18 chitinase that releases monomers during substrate hydrolysis. Microbiology. 2012;158(8):2168–2179.
- Velmurugan N, Kalpana D, Han JH, et al. A novel low temperature chitinase from the marine fungus Plectosphaerella sp. strain MF-1. Botanica Marina. 2011;54(1):75–81.
- Yang S, Fu X, Yan Q, et al. Biochemical characterization of a novel acidic exochitinase from Rhizomucor miehei with antifungal activity. J Agric Food Chem. 2016;64(2):461–469.
- Yu G, Xie LQ, Li JT, et al. Isolation, partial characterization, and cloning of an extracellular chitinase from the entomopathogenic fungus Verticillium lecanii. Genet Mol Res. 2015;14(1):2275–2289.
- Deane EE, Whipps JM, Lynch JM, et al. The purification and characterization of a Trichoderma harzianum exochitinase. Biochim Biophys Acta. 1998;1383(1):101–110.
- Kumar DP, Singh RK, Anupama PD, et al. Studies on exo-chitinase production from Trichoderma asperellum UTP-16 and its characterization. Ind J Microbiol. 2012;52(3):388–395.
- Sahai AS, Manocha MS. Chitinases of fungi and plants: their involvement in morphogenesis and host-parasite interaction. FEMS Microbiol Rev. 1993;11(4):317–338.
- Hartl L, Zach S, Seidl-Seiboth V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol. 2012;93(2):533–543.
- Sood M, Kapoor D, Kumar V, et al. Trichoderma: the “secrets” of a multitalented biocontrol agent. Plants. 2020;9(6):762.
