Abstract
Penicillium species have been actively studied in various fields, and many new and unrecorded species continue to be reported in Korea. Moreover, unidentified and misidentified Korean Penicillium species still exist in GenBank. Therefore, it is necessary to revise the Korean Penicillium inventory based on accurate identification. We collected Korean Penicillium nucleotide sequence records from GenBank using the newly developed software, GenMine, and re-identified Korean Penicillium based on the maximum likelihood trees. A total of 1681 Korean Penicillium GenBank nucleotide sequence records were collected from GenBank. In these records, 1208 strains with four major genes (Internal Transcribed Spacer rDNA region, β-tubulin, Calmodulin and RNA polymerase II) were selected for Penicillium re-identification. Among 1208 strains, 927 were identified, 82 were identified as other genera, the rest remained undetermined due to low phylogenetic resolution. Identified strains consisted of 206 Penicillium species, including 156 recorded species and 50 new species candidates. However, 37 species recorded in the national list of species in Korea were not found in GenBank. Further studies on the presence or absence of these species are required through literature investigation, additional sampling, and sequencing. Our study can be the basis for updating the Korean Penicillium inventory.
1. Introduction
Penicillium species have been widely studied in various fields including pharmacy, industry, and taxonomy [Citation1–4]. Species with different properties are used in Penicillium studies, and correct identification of Penicillium species is important for scientific communication and verification. It is difficult to identify Penicillium at the species level because of intra-species variations and similar morphological characteristics among species [Citation5]. Therefore, DNA sequence-based identification was introduced, which dramatically progressed the taxonomy of Penicillium. In fungal taxonomy, the internal transcribed spacer (ITS) rDNA region is used as a general DNA barcode marker in kingdom fungi [Citation6]. Although ITS sequences can be used to identify Penicillium sections and certain species, they have insufficient resolution for precise identification of closely related Penicillium species. β-tubulin (BenA) has been recommended as a secondary marker for the identification of Penicillium species due to ease of amplification, sufficient resolution among closely related species, and high coverage on existing Penicillium species [Citation5]. The combined phylogeny of ITS, BenA, calmodulin (CaM) and RNA polymerase II (RPB2) datasets has been proposed to describe new species [Citation5].
Sequence databases are essential resources for molecular species identification in taxonomy. GenBank is currently the largest and most widely used public sequence repository provided by the National Center for Biotechnology Information (NCBI) and a division of the National Library of Medicine (NLM), which includes more than 170,000 Penicillium nucleotide sequences [Citation7]. The advantage of using GenBank as a reference database for molecular taxonomic studies is that it enables the comparison of sample data with comprehensive sequences from around the world [Citation8]. BLAST (Basic Local Alignment Search Tool) search on GenBank databases has been frequently used as a method for Penicillium species identification because of its easy accessibility and rapid generation of results. However, an open access submission system of GenBank had led GenBank to include invalid sequences such as misidentified and wrongly edited sequences, which resulted in incorrect identifications in taxonomic studies using BLAST [Citation9,Citation10]. Therefore, a precisely curated sequence database for Penicillium is required.
Many studies have been conducted recently on Penicillium species in Korea. Numerous new and unrecorded species have been reported from marine and terrestrial environments continuously over a period of time [Citation1,Citation2,Citation11,Citation12]. To date, 117 Penicillium species have been reported in Korea [Citation13]. Some Penicillium species in the previous Korean Penicillium inventory were identified based only on morphology or ITS sequence [Citation13,Citation14]. In addition, a number of Korean Penicillium records in GenBank were misidentified because of the sole reliance on the ITS sequence. Therefore, the previous Korean Penicillium inventory needs to be revised for precise identification. In this study, we intended to re-identify Korean Penicillium in GenBank based on the maximum likelihood (ML) tree and build a robust Korean inventory based on these results.
2. Materials and methods
2.1. Collection of GenBank records
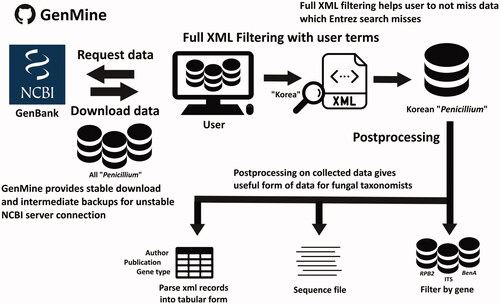
GenBank records of Korean Penicillium sequences were collected from the GenBank nucleotide database using the web-crawling method and filtered by gene names (i.e., term “internal transcribed spacer,” “calmodulin,” etc.) in sequence descriptions. Non-Korean records (i.e., Canadian Penicillium study with Korean authors) or non-fungal records (i.e., Penicillium virus sequences) collected by automatic word match were manually inspected and excluded through author and publication information provided by the GenMine software. Metadata of composition and annual record increment for each gene of Korean Penicillium records were analyzed and visualized. Collected records were merged for phylogenetic analysis using strain names to avoid duplicate sequences from the same specimen.
Python scripts used for GenBank record collection and post-processing in this research has been modified and generalized to a standalone software, GenMine (). It provides stable data crawling from GenBank, term search in comprehensive raw xml GenBank record data, sorting by gene names, and tabular data transformation from raw records. GenMine is available at https://github.com/Changwanseo/GenMine in GPL-3.0 License.
2.2. Re-identification of Korean Penicillium in GenBank
Sequences from type species of Penicillium were used as a reference dataset for identification [Citation15]. ITS sequences cross-validated with correlated protein coding genes were used for additional reference dataset. The ITS, BenA, CaM, and RPB2 sequences of Korean Penicillium strains from GenBank were aligned with reference sequences using MAFFT 7.453 software with the L-INS-i algorithm (–localpair), 1000 iterative refinement cycles (–maxiterate 1000), gap opening penalty 1.3 (–op 1.3), and gap extension penalty 0.1 (–ep 0.1) options, respectively [Citation16]. Manual end trimming and alignment editing were performed on the aligned sequences to remove non-informative regions. ML trees were built with aligned and trimmed sequences using RAxML 8.2.12. in rapid bootstrap analysis, and the best-scoring ML tree was found with the GTRGAMMA model (-m GTRGAMMA) and 1000 bootstrap replicates (-# 1000) options [Citation17]. Korean Penicillium sequences were re-identified based on the branch lengths and bootstrap values of the ML trees (Figure S1–S4). Clades with branch support over 70% of bootstraps, and branch length over 0.002(ITS) and 0.01(BenA, CaM, RPB2) were generally treated as clearly distinct branches. For Penicillium species with narrower or boarder species ranges than general standards, interpretation of original publications of the corresponding species was applied for identification. The final re-identification results were organized to Korean Penicillium inventory. The overall re-identification workflow is illustrated.
3. Results
3.1. Composition and growth of Korean Penicillium records in GenBank
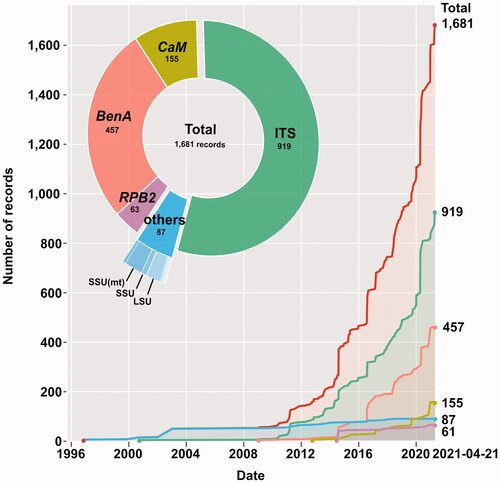
A total of 1681 Korean Penicillium sequences with 175 assigned species were collected from the GenBank nucleotide database on April 21st, 2021 (; Table S5). The collected sequences consisted of 919 ITS, 457 BenA, 155 CaM, 63 RPB2, and 59 others. The others included taxonomic genes (18S ribosomal small subunit (SSU), 28S ribosomal large subunit (LSU), mitochondrial small subunit, elongation factor 1-α (ef1-α), and cytochrome oxidase subunit 1(cox1)), and genes for genetic studies (chitin synthase (chs1, chs2, chs3, and chs4), α-L-arabinofuranosidase (abfB), endo-xylanase (xynY), phytase PJ3, mitochondrial rRNA, and tRNA genes). The first Korean Penicillium sequence records were chitin synthase genes (U57321, U57322, U57323), deposited in 1996 (). The first ITS Korean Penicillium sequence record was deposited in GenBank in 2000, which was misidentified as Paecilomyces in the initial submission (AF291869) [Citation18]. Apart from this, the currently assigned Korean Penicillium ITS sequences have been continuously deposited since 2008. Among protein-coding genes, BenA sequences have been deposited from 2009, and deposits of CaM and RPB2 sequence records started in 2014. A total of 1594 sequences of four major genes (ITS, BenA, CaM, and RPB2) of Penicillium taxonomy were used for re-identification. These sequences were from 83 publications (Table S1). In terms of strains, 742 strains had only ITS sequence, 280 strains had BenA sequence but no ITS sequence, and 172 strains had both ITS and BenA sequences.
3.2. Re-identification of Korean Penicillium in GenBank
Re-identification of 742 strains with only ITS sequence resulted in 385 correctly identified strains (86 spp.), 74 strains of new species candidates (30 spp.), and 202 strains that were incapable of identification (). The remaining 81 strains were identified as other genera, including Talaromyces (n = 76), Cladosporium (n = 2), Aureobasidium (n = 1), and unidentified genera (n = 2). The 742 ITS-only strains consisted of 389 strains assigned at the species level and 353 strains unassigned. Among the 389 identified assigned strains, 146 were found to be correctly identified and 243 were misidentified.
Figure 3. Diagram of the re-identification process and results on Korean Penicillium ITS and BenA sequences from GenBank. The sequences were re-identified by RAxML tree-based identification and compared with original annotation of corresponding GenBank record. Sequences without scientific names is GenBank record were labeled “Unassigned,” and sequences cannot be identified due to low resolution of phylogenetic tree were labeled as “Undetermined.” Numbers in the diagram show the number of records in each category.
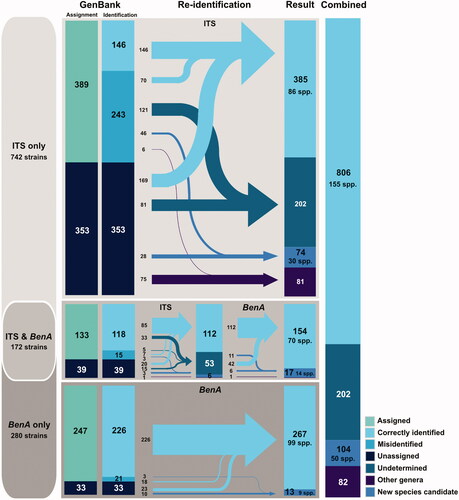
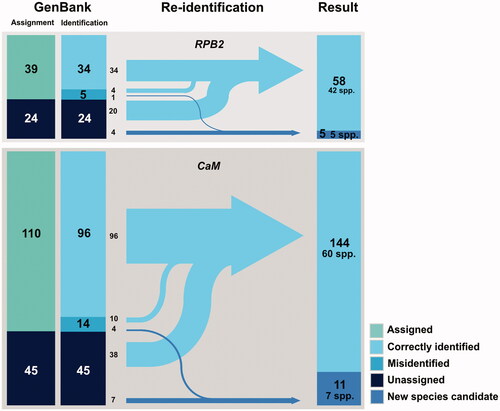
Re-identification of 280 strains with BenA sequences that had no corresponding sequences of ITS sequences resulted in 267 identified strains (99 spp.) and 13 new species candidate strains (9 spp.). The 280 strains consisted of 247 strains assigned at the species level and 33 strains unassigned. There were 21 (out of 247) strains that were misidentified. Of the 172 strains with both ITS and BenA sequences, re-identification resulted in 154 correctly identified strains (70 spp.), 17 new species candidate strains (14 spp.), and one Talaromyces. Among 133 strains assigned at the species level within the 172 strains with both ITS and BenA, 118 were correctly identified, and 15 turned out to be misidentified. For the 155 strains with CaM sequences, there were 144 correctly identified strains (60 spp.) and 11 new species candidate strains (7 spp.) (). Among 110 strains assigned at the species level within the 155 strains, 96 turned out to be correctly identified and 14 were misidentified. For the 63 strains with RPB2 sequences, 58 were correctly identified (42 spp.) and 5 were new species candidate strains (5 spp.). Among 39 strains assigned at the species level, 34 strains were correctly identified. All re-identification results are listed in Table S6.
Figure 4. Diagram of the re-identification process and results of Korean Penicillium RPB2 and CaM sequences from GenBank. The sequences were re-identified by RAxML tree-based identification and compared with original annotation of corresponding GenBank record. Sequences without scientific names is GenBank record were labeled “Unassigned,” and sequences cannot be identified due to low resolution of phylogenetic tree were labeled as “Undetermined.” Numbers in the diagram show the number of records in each category.
4. Discussion
We identified 927 Korean Penicillium strains from 1594 sequences (1208 strains) in GenBank to establish an accurate Korean Penicillium inventory (Table S2). As a result, approximately 206 Penicillium species are predicted to exist in Korea: 156 recorded species and 50 new species candidates. Compared to the national list of species in Korea [Citation13], 80 species were previously recorded, and 76 species were unrecorded in Korea. However, 37 species recorded in national list of species of Korea were not observed in this study ().
Table 1. Number of Korean Penicillium strains and species found in GenBank.
ITS-based identification constitutes the majority of Penicillium identification in Korea [Citation19–21]. According to our results on Korean Penicillium species with ITS sequences, 69.6% could be identified solely by their ITS sequence with ML tree-based identification (Table S3). However, even though ITS is a universal taxonomic marker for fungi, it is insufficient for the precise identification of Penicillium species due to its low resolution [Citation6]. Our results showed that 62.5% of Penicillium strains identified based solely on ITS were misidentified, and 27.2% (202 out of 742) remained undetermined due to indistinguishable tree topology. Protein-coding genes were widely used in the identification of Penicillium species in Korea by Park et al. in 2014 assisted by the Marine Fungal Resource Bank (MFRB) by the Ministry of Maritime Affairs and Fisheries [Citation1]. Our results showed an accuracy of 91.4% when identification was based solely on BenA sequence, and 88.7% when ITS and BenA were combined. Although the sample size is relatively small, identification accuracies of RPB2 and CaM showed 87.2% and 87.3% In addition, all records can be taxonomically re-assigned at the species level after re-identification with protein-coding genes. Therefore, as the BenA sequence is a precise marker with abundant database in Penicillium species, Penicillium researchers should use ITS and BenA sequences and other secondary markers such as CaM or RPB2 for precise identification [Citation5].
GenBank has enabled comprehensive studies of sequences produced worldwide. However, despite the advantages in generality and accessibility, precautions are required when using GenBank for identification based on the evidence of cases observed in this study. First, GenBank contains misidentified sequences due to open access system [Citation22–25]. Our results showed that 32.5% of the Korean Penicillium sequences were misidentified. These misidentified sequences could mislead researchers into false identification if the sequences were used as reference dataset. Therefore, it is important to double check reference data from multiple resources and use reference verifiable methods such as the ML tree. Second, GenBank includes sequences with incorrect edits [Citation26,Citation27]. We observed that in certain cases, ITS and BenA sequences from the same strain were identified as belonging to different species in (MH374608 and MH367044). Due to unusual variations in the conserved region of the ITS sequence, we hypothesized that the misidentification was derived from incorrect editing. The quality of the GenBank records cannot be verified because of the absence of raw chromatograms, and users should recognize that GenBank records might contain errors. Finally, the GenBank search result does not guarantee completeness because of omitted metadata tags and deficiency of default search system of GenBank, Entrez [Citation28]. With GenMine, we observed that several records were absent in the Entrez search results. Therefore, sequence uploaders should provide Darwin Core formatted metadata, and researchers should be aware that Entrez search results are not complete [Citation29].
GenMine has played role for collecting and primary analysis for Penicillium sequence data in this analysis. APIs like Entrez and data collection software such as SUPERCRUNCH were developed previously for GenBank data collection [Citation28,Citation30]. However, most of them were for genomic research or low-level APIs, and none of them were for taxonomists for comprehensive study of fungal barcode genes. GenMine can collect sequences with tags that cannot be searched by Entrez term system by scheming full xml records. GenMine provides useful features for taxonomists such as gene classification, author and journal information extraction, xml record tabulating, and internet stability for large data in single command. GenMine can be useful tool for fungal taxonomists to analyze other fungal taxa in GenBank and improve misidentifications in GenBank.
At the early stage of the sequence-based Korean Penicillium study, a majority of the strains were isolated from the soil environment and focused on morphology for diversity studies [Citation14]. Recently, isolation sources have branched out to various environments including marine, freshwater, indoor, and air [Citation31–34]. New and unrecorded species in Korea have increased dramatically [Citation1,Citation12,Citation13,Citation35,Citation36]. Further studies on the presence or absence of these species recorded only in the national list of species of Korea are required through literature investigation, additional sampling, and sequencing. In addition, for these unrecorded species and new species candidates, it is necessary to update the inventory of Korea through comprehensive additional research on morphology and sequence. Our study could serve as a basis for updating the Korean Penicillium inventory. Furthermore, this study could contribute to Penicillium studies by providing results for adjusting misidentifications of Penicillium strains in GenBank.
Supplemental Material
Download MS Excel (301.3 KB)Supplemental Material
Download MS Excel (279.2 KB)Supplemental Material
Download MS Excel (10.2 KB)Supplemental Material
Download MS Excel (9.7 KB)Supplemental Material
Download MS Excel (56.9 KB)Supplemental Material
Download MS Excel (19 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Park MS, Fong JJ, Oh S-Y, et al. Marine-derived Penicillium in Korea: diversity, enzyme activity, and antifungal properties. Antonie Van Leeuwenhoek. 2014;106(2):331–345.
- Nguyen TT, Pangging M, Bangash NK, et al. Five new records of the family Aspergillaceae in Korea, Aspergillus europaeus, A. pragensis, A. tennesseensis, Penicillium fluviserpens, and P. scabrosum. Mycobiology. 2020;48(2):81–94.
- He F, Li X, Yu J-H, et al. Secondary metabolites from the mangrove sediment-derived fungus Penicillium pinophilum SCAU037. Fitoterapia. 2019;136:104177.
- Honary S, Barabadi H, Gharaei-Fathabad E, et al. Green synthesis of copper oxide nanoparticles using Penicillium aurantiogriseum, Penicillium citrinum and Penicillium waksmanii. Dig J Nanomater Bios. 2012;7(3):999–1005.
- Visagie C, Houbraken J, Frisvad JC, et al. Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014;78:343–371.
- Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246.
- Sayers EW, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2021;49(D1):D92–D96.
- Meiklejohn KA, Damaso N, Robertson JM. Assessment of BOLD and GenBank–their accuracy and reliability for the identification of biological materials. PLoS One. 2019;14(6):e0217084.
- Lee YG, Chung K-C, Wi SG, et al. Purification and properties of a chitinase from Penicillium sp. LYG 0704. Protein Expr Purif. 2009;65(2):244–250.
- Ngan NTT, Quang TH, Kim K-W, et al. Anti-inflammatory effects of secondary metabolites isolated from the marine-derived fungal strain Penicillium sp. SF-5629. Arch Pharm Res. 2017;40(3):328–337.
- Kim WK, Sang HK, Woo SK, et al. Six species of Penicillium associated with blue mold of grape. Mycobiology. 2007;35(4):180–185.
- Park MS, Lee S, Lim YW. A new record of four Penicillium species isolated from Agarum clathratum in Korea. J Microbiol. 2017;55(4):237–246.
- National List of Species of Korea. 2020. National Institute of Biological Resources. [cited 2021-Sep 30]. Available from: http://kbr.go.kr.
- Lee S, Hong S-B, Kim C-Y. Contribution to the checklist of soil-inhabiting fungi in Korea. Mycobiology. 2003;31(1):9–18.
- Houbraken J, Kocsubé S, Visagie C, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313.
- Park J-E, Kim G-Y, Park H-S, et al. Phylogenetic analysis of caterpillar fungi by comparing ITS1-5.8S-ITS2 ribosomal DNA sequences. Mycobiology. 2001;29(3):121–131.
- Khan SA, Hamayun M, Yoon H, et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008;8(1):231–210.
- Kim CS, Park MS, Yu SH. Two species of endophytic Penicillium from Pinus rigida in Korea. Mycobiology. 2008;36(4):222–227.
- Bae K-S, Hong S-G, Park Y-D, et al. Sequence comparison of mitochondrial small subunit ribosomal DNA in Penicillium. Journal of Microbiology. 2000;38(2):62–65.
- Lücking R, Aime MC, Robbertse B, et al. Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 2020;11(1):1–32.
- Jung PE, Fong JJ, Park MS, et al. Sequence validation for the identification of the white-rot fungi Bjerkandera in public sequence databases. J Microbiol Biotechnol. 2014;24(10):1301–1307.
- Jargalmaa S, Eimes JA, Park MS, et al. Taxonomic evaluation of selected Ganoderma species and database sequence validation. PeerJ. 2017;5:e3596.
- Pentinsaari M, Ratnasingham S, Miller SE, et al. BOLD and GenBank revisited–do identification errors arise in the lab or in the sequence libraries? PLoS One. 2020;15(4):e0231814.
- Buhay JE. “COI-like” sequences are becoming problematic in molecular systematic and DNA barcoding studies. J Crustacean Biol. 2009;29(1):96–110.
- Fietz K, Graves JA, Olsen MT. Control control control: a reassessment and comparison of GenBank and chromatogram mtDNA sequence variation in Baltic grey seals (Halichoerus grypus). PLoS One. 2013;8(8):e72853.
- Schuler GD, Epstein JA, Ohkawa H, et al. [10] Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162.
- Aime MC, Miller AN, Aoki T, et al. How to publish a new fungal species, or name, version 3.0. IMA Fungus. 2021;12(1):1–15.
- Portik DM, Wiens JJ. SuperCRUNCH: a bioinformatics toolkit for creating and manipulating supermatrices and other large phylogenetic datasets. Methods Ecol Evol. 2020;11(6):763–772.
- Ha TM, Ko W, Lee SJ, et al. Anti-inflammatory effects of curvularin-type metabolites from a marine-derived fungal strain Penicillium sp. SF-5859 in lipopolysaccharide-induced RAW264. 7 Macrophages. Marine Drugs. 2017;15(9):282.
- Heo I, Hong K, Yang H, et al. Diversity of Aspergillus, Penicillium, and Talaromyces species isolated from freshwater environments in Korea. Mycobiology. 2019;47(1):12–19.
- Oh J-Y, Kim E-N, Ryoo M-I, et al. Morphological and molecular identification of Penicillium islandicum isolate KU101 from stored rice. Plant Pathol J. 2008;24(4):469–473.
- Kim KY, Kim CN. Airborne microbiological characteristics in public buildings of Korea. Build Environ. 2007;42(5):2188–2196.
- Park MS, Fong JJ, Oh S-Y, et al. Penicillium jejuense sp. nov., isolated from the marine environments of Jeju Island, Korea. Mycologia. 2015;107(1):209–216.
- Park MS, Lee EJ, Fong JJ, et al. A new record of Penicillium antarcticum from marine environments in Korea. Mycobiology. 2014;42(2):109–113.