Abstract
Two strains, YP344 and YP579 were isolated from soil samples in Pocheon City, Gyeonggi Province, South Korea. The strains YP344 and YP579 belong to the genus Vishniacozyma and Dioszegia, respectively. The molecular phylogenetic analysis showed that the strain YP344 was closely related to Vishniacozyma peneaus. Strain YP344T differed by four nucleotide substitutions with no gap (0.70%) in the D1/D2 domain of the LSU rRNA gene and 16 nucleotide substitutions with 8 gaps (5.76%) in the ITS region. On the other hand, the strain YP579T varied from the type strain of the most closely related species, Dioszegia zsoltii var. zsoltii, by 6 nucleotide substitutions with four gaps (1.64%) in the D1/D2 domain of LSU rRNA gene and 26 nucleotide substitutions with 14 gaps (8.16%) in the ITS region. Therefore, the name Vishniacozyma terrae sp. nov. and Dioszegia terrae sp. nov. are proposed, with type strains YP344T (KCTC27988T) and YP579T (KCTC 27998T), respectively.
1. Introduction
The yeast belonging to the basidiomycetous are currently recognized in three classes of the phylum Basidiomycota: Ustilaginomycetes, Urediniomycetes, and Hymenomycetes [Citation1]. These yeasts belong to those phyla that have considerable agricultural, medical, and economic importance. An estimate suggested that only up to 5% of the yeast species belong to the Basidiomycota in nature. At the time of writing (October 2022), about 500 species of basidiomycetous yeasts have been widely recognized; among them, the psychrophilic yeast belonging to the genus Vishniacozyma and Rhodotorula have been discovered in northern regions, glacial mountains, and polar habitats [Citation2–4]. The fungi that adapted to grow in the cold environment can decompose diverse types of organic compounds that play an essential role in the nutrient cycles of polar microbial ecosystems [Citation5–8].
Dioszegia genus forms a monophyletic group within the Tremellales (Tremellomycetes, Agaricomycotina) [Citation9,Citation10]. In 2015, Liu et al. [Citation9] reconstructed the phylogeny of the tremellomycetous yeasts and related dimorphic and filamentous Tremellomycetes using sequence analysis, and proposed a novel family Bulleribasidiaceae for Dioszegia within the order Tremellales. As of this writing, all species characterized so far are nonfermentative, may or may not form ballistoconidia, and do not have evidence of a sexual stage. Most species of Dioszegia have been isolated from leaves, roots, or soil [Citation11]. There is an accumulation of carotenoid pigments within the cells of yeasts in the genus Dioszegia, resulting in the salmon, pink or red color of their colonies [Citation12,Citation13].
As part of our yeast biodiversity study in Pocheon, South Korea, we isolated three strains of the genus Vishniacozyma and one strain of the genus Dioszegia from a soil sample. According to sequence analysis of the D1/D2 domain of the large subunit rRNA gene and the internal transcribed spacer (ITS) region, these yeasts are closely related to Vishniacozyma peneaus and Dioszegia zsoltii var. zsoltii, in the Tremellales, Tremellomycetes, and Basidiomycota.
2. Materials and methods
2.1. Yeast isolation and phenotypic characteristics
The soil samples were collected in Pocheon city, Gyeonggi Province, South Korea (37°46′18.7″N 127°09′46.3″E, 37°51′19.2″N 127°10′56.6″E, 37°50′38.4″N 127°09′34.1″E, and 37°47′54.8″N 127°10′01.6″E) during winter (). The soil sample (1 g) was suspended in 9 ml of sterile saline and serially diluted until they were 1:10 to 1:1000, and then spread 0.1 ml of each on Yeast-Malt Agar (YMA, Difco, Detroit, USA) plates and incubated at 25 °C for 3–4 days. The strains YP344, YP155, YP333, and YP579 were isolated and purified by cross-streaking on a YM agar medium (1% yeast extract, 2% peptone, 2% glucose, and 1.5% agar). The pure culture of these strains was deposited at the Korea Collection for Type Cultures, KRIBB, Korea. The purified yeasts were maintained in 20% v/v glycerol at −80 °C.
Table 1. List of the yeast strains of Vishniacozyma terrae sp. nov. examined in the present study and related species.
2.2. DNA sequencing and phylogenetic analysis
The four yeast strains (YP344T, YP155, YP333, and YP579T) were identified by analysis of D1/D2 domain of the LSU rRNA gene and ITS region. The genomic DNA was amplified by PCR with NL1/NL4 [Citation14] and ITS1/ITS4 primers [Citation15], respectively. In order to assemble the contigs, the SeqMan program version 7.1.0 was used. The BLAST search of pairwise sequences [Citation16] was conducted and alignments were performed with Clustal X 2.0 [Citation17] to match sequences from related species. Based on the combined sequences of the ITS region and the D1/D2 domains of the LSU rRNA gene, phylogenetic trees were constructed using the MEGA X program [Citation18–20]. The evolutionary distances were calculated by the general time reversible (GTR) and Kimura two-parameter model for the maximum-likelihood and neighbor-joining analyses, respectively [Citation21,Citation22]. A bootstrap analysis was also conducted using 1000 replicates [Citation23]. The BLAST tool software (https://blast.ncbi.nlm.nih.gov), was used to calculate sequence similarities and nucleotide substitutions in ITS and D1/D2 regions between newly isolated strains and closely related species.
2.3. Phenotypic characterization
For the microscopic examination, four strains were cultivated on YM agar at 15 °C and examined using a phase-contrast microscope (DM500, LEICA, Wetzlar, Germany). The strains were phenotypically characterized by the methods reported by Kurtzman et al. [Citation24]. For the physical examination, strains were cultivated on YM agar at 15 °C and examined with a phase-contrast microscope (DM500). To stimulate the sexual reproductive phase and spore formation of the yeast cells, single or mixed cultures of the both strains were incubated at 10 °C for 2 months on YM agar. Pseudohyphae and true hyphae have been observed weekly during cell culture on YM agar at 10 °C for up to 2 months. Basidiospore formation was confirmed by growing the individual strains on corn meal agar (2% corn meal infusion and 2% agar), 5 % malt extract agar (5 % malt extract and 1.5 % agar), potato dextrose agar (PDA, Difco, Detroit, USA), YM agar, and yeast extract–peptone glucose (YPD, Difco, Detroit, USA) at 25 °C for 2 months. The color reaction with diazonium blue B (DBB, Sigma-Aldrich, Darmstadt, Germany) was performed by dropping the DBB reagent into the 3 days incubated colonies grown on YM agar medium and observing the color development after 2 min. Growth was accessed at various temperatures (4 °C, 10 °C, 15 °C, 25 °C, 30 °C, 35 °C, 37 °C, 42 °C, and 45 °C) and determined by cultivation on PDA, YPD agar, and YMA for 15 days. Growth in YM broth with different NaCl concentrations (0%–10% in 1% intervals, w/v) was studied for 1–5 days. Ubiquinone was prepared, extracted and analyzed as described by Prillinger et al. [Citation25].
3. Results and discussion
3.1. Species identification and delineation
In the study, a total of 472 yeast strains were isolated from 35 soil samples collected in the city of Pocheon, Gyeonggi Province, South Korea. Of these strains, 11 were classified as Vishniacozyma (taxonomy: Basidiomycota, Agaricomycotina, Tremellomycetes, Tremellales, Bulleribasidiaceae) and one were classified as Dioszegia (taxonomy: Basidiomycota, Agaricomycotina, Tremellomycetes, Tremellales, Bulleribasidiaceae) by analyzing sequences of the ITS and the D1/D2 domain of the LSU rRNA gene. As a result, three strains (YP344, YP155 and YP333) were classified as the new Vishniacozyma species and one strain (YP579) was classified as the new Dioszegia species. MycoBank numbers of the strains YP344 and YP579 are MB841262 and MB841263, respectively.
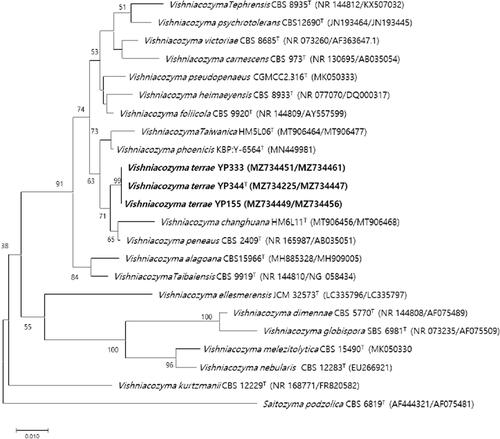
Strain YP344 was most closely related to V. peneaus with 94.2% sequence identity, against which 4 nt substitutions were observed in the D1/D2 domains and 16 nt substitutions in the ITS regions of YP344 were replaced by 16 nt substitutions. Phylogenetic trees constructed by maximum-likelihood and neighbor-joining methods showed that strains YP344, YP155 and YP333 were grouped with members of the genus Vishniacozyma ( and Supplementary Figure S1).
Figure 1. Phylogenetic tree based on the concatenated sequences of the D1/D2 region of the LSU rRNA gene and ITS regions and constructed by the neighbor-joining method, shows relationships between strains of a novel species (YP344, YP155, and YP333) and closely related species. The novel species described in this manuscript are highlighted in bold. Saitozyma podzolica CBS 6819T was used as outgroup. Bootstrap values >50% (% of 1000 replications) were shown at branch points. Accession numbers were shown in parentheses. Bar, 0.01 substitutions per nucleotide position.
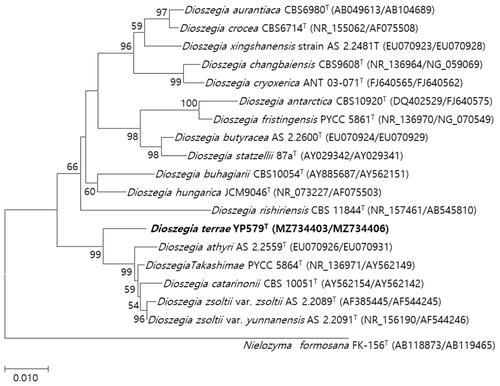
The strain YP579 was most closely related to D. zsoltii var. zsoltii with sequence identities of 91.84%, against which 6 nt substitutions were observed in the D1/D2 domain () and 26 nt the sequence of the ITS region. The phylogenetic trees generated by maximum-likelihood and neighbor-joining methods showed that strain YP579 was grouped with members of the genus Dioszegia ( and Supplementary Figure S1). The type strain YP344T did not assimilate glycerol and could not grow at 10% NaCl but V. peneaus assimilated glycerol and was able to grow at 10% NaCl ( and ). Ballistoconidia are produced on YM agar. Basidiospore formation is not observed on corn meal agar, PDA, YPD agar, 5 % malt extract agar, and YM agar at 15 and 25 °C for 2 months. The respiratory quinone is Q-10.
Table 2. List of the yeast strains of Dioszegia terrae sp. nov. examined in the present study and related species.
Table 3. Nucleotide substitutions in the sequences of the D1/D2 domain of the LSU rRNA gene and ITS region of Vishniacozyma terrae sp. nov. (YP344T) and Vishiniacozyma species.
Table 4. Nucleotide substitutions in the sequences of the D1/D2 domain of the LSU rRNA gene and ITS region of Dioszegia terrae sp. nov. (YP579T) and Dioszegia species.
Diazonium blue B and urease reactions are positive, consistent with the characteristics of the genus Dioszegia [Citation24,Citation26]. In addition, the type strain YP579 did not assimilate galactitol and d-mannitol. In contrast, D. zsoltii var. zsoltii did assimilate galactitol and d-mannitol. Ballistoconidia are produced on YM agar after 53 days of incubation. Basidiospore formation is not observed on PDA, corn meal agar, 5 % malt extract agar, YPD agar, and YM agar at 15 and 25 °C for 2 months. The respiratory quinone is Q-10. Based on these results, YP344 and YP579 should be considered a new species, for which the name Vishniacozyma terrae (terrae of the soil) and Dioszegia terrae (terrae of the soil) is proposed.
3.2. Description of V. terrae Park, Maeng, and Sathiyaraj sp. nov.
V. terrae (ter′rae. L. gen. n. terrae of the soil, referring to the isolation source of the type strain)
Novel yeast species belonging to phylum Basidiomycota, subphylum Agaricomycotina, class Tremellomycetes, order Tremellales, family Bulleribasidiaceae.
Yeast cells after three days on YM agar at 15 °C are ovoid (6–6.5 × 1.8–2 μm). Budding is polar budding (). Streak culture on YM agar for 1 week at 15 °C produces cream–color, convex, round, shiny, and slimy colonies. Pseudohyphae and true hyphae are not formed after 53 days of incubation on YMA, PDA, and CMA. Ballistoconidia are not formed after 53 days of incubation on YMA, PDA, and CMA. Basidiospore formation is not formed after 53 days of incubation on YMA, PDA, and CMA.
Figure 2. Phylogenetic tree based on the concatenated sequences of the D1/D2 region of the LSU rRNA gene and ITS regions and constructed by the maximum-likelihood method, shows relationships between strains of a novel species (YP579T) and closely related species. The novel species described in this manuscript are highlighted in bold. Nielozyma formosana FK-156T was used as outgroup. Bootstrap values >50% (% of 1000 replications) were shown at branch points. Accession numbers were shown in parentheses. Bar, 0.01 substitutions per nucleotide position.
Glucose, 2-keto-d-gluconate, l-arabinose, d-xylose, d-galactose, inositol, d-sorbitol, N-acetyl-d-glucosamine, d-cellobiose, d-lactose (bovine origin), d-maltose, d-saccharose (sucrose), d-melezitose, adonitol, d-trehalose, and d-raffinose are assimilated, but glycerol, xylitol, and d-methyl-d-glucoside are not assimilated. Growth occurs at 15 °C–37 °C (optimum 25 °C) and cells can tolerate up to 6% NaCl in YM broth. Growth occurs on YM agar, YPD agar, and 50% glucose medium. Growth with 0.01% of cycloheximide is positive. Production of starch and diazonium blue B reaction and urea hydrolysis are negative. The respiratory quinone is Q-10 ( and ).
Table 5. Phenotypic characteristics that differentiate Vishniacozyma terrae sp. nov. and related species, V. peneaus and V. phoenicis.
Table 6. Phenotypic characteristics that differentiate Dioszegia terrae sp. nov. and related species, D. zsoltii and D. catarinonii.
The holotype, YP344T, was isolated from the soil sample in Pocheon city, Gyeonggi Province in Korea, and is preserved in a metabolically inactive state at the Korea Collection for Type Cultures, KRIBB, Korea as KCTC 27988T. The GenBank accession numbers for the D1/D2 domain of the LSU rRNA gene and ITS region for YP344T are MZ734225 and MZ734447, respectively. The MycoBank accession number is MB 841262.
3.3. Description of D. terrae Park, Maeng, and Sathiyaraj sp. nov.
D. terrae (ter′rae. L. gen. n. terrae of the soil, referring to the isolation source of the type strain)
Novel yeast species belonging to phylum Basidiomycota, subphylum Agaricomycotina, class Tremellomycetes, order Tremellales, family Bulleribasidiaceae.
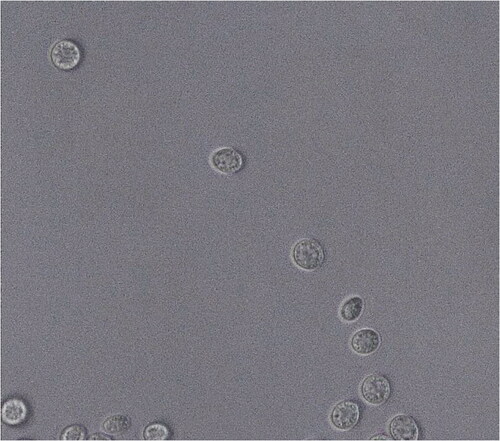
Yeast cells after 3 days on YM agar at 15 °C are ovoid (6–6.5 × 1.8–2 μm). Budding is polar budding ( and ). Pseudohyphae and true hyphae are not formed after 53 days of incubation. Streak culture are on YM agar for 1 week at 15 °C and produces colonies that are orange–color, convex, round, and slimy. Ballistoconidia is formed after 53 days of incubation but basidiospore is not formed ().
Figure 3. Vishniacozyma terrae sp. nov. YP344T and Dioszegia terrae sp. nov. YP579T (A) The polar budding cells of V. terrae YP344T and (B) D. terrae YP579T on YM agar after 3 days at 10 °C.
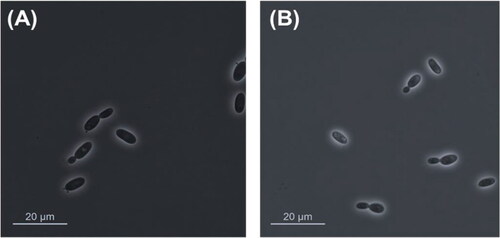
Figure 4. Dioszegia terrae sp. nov. YP579T ballistoconidia produced on YM agar after 53 days at 10 °C. Bars, 10 μm.
Glucose, sucrose, raffinose, melibiose, galactose, trehalose, maltose, melezitose, soluble starch, cellobiose, l-rhamnose, d-xylose, l-arabinose, d-arabinose, d-ribose, xylitol, d-gluconate, d-glucosamine, N-acetyl-d-glucosamine, potassium nitrate, cadaverine dihydrochloride, and l-lysine are assimilated. Inulin, lactose, methyl-a-d-glucoside, l-sorbose, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, d-mannitol, d-glucitol, myo-inositol, dl-lactate, citrate, gluconolactone, and sodium nitrate are not assimilated.
Growth occurs at 10 °C–30 °C (optimum 18 °C) and cells can tolerate up to 4% NaCl in YM broth. Growth occurs on YM agar, YPD agar, and PDA medium but not on 50% glucose medium. Growth in the presence of 0.01% of cycloheximide is positive. Production of starch and diazonium blue B reaction and urea hydrolysis are positive. The respiratory quinone is Q-10.
The holotype, YP579T, was isolated from the soil sample in Pocheon city, Gyeonggi province, South Korea, and is preserved in a metabolically inactive state at the Korea Collection for Type Cultures, KRIBB, Korea as KCTC 27998T. The GenBank/EMBL/DDBJ accession numbers for the D1/D2 domain of the LSU rRNA gene and ITS region for YP579T are MZ734403 and MZ734406, respectively. The MycoBank accession number is MB 841263.
Supplemental Material
Download MS Word (73 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kurtzman CP, Fell JW, Boekhout T. The yeasts. 5th ed. Burlington: Elsevier; 2011. p. 1339–1372.
- Sibanda EP, Mabandla M, Chisango T, et al. Endophytic fungi isolated from the medicinal plants Kigelia africana and Warburgia salutaris. CBIOT. 2018;7(4):323–328.
- Ogaki MB, Teixeira DR, Vieira R, et al. Diversity and bioprospecting of cultivable fungal assemblages in sediments of Lakes in the antarctic peninsula. Fungal Biol. 2020;124(6):601–611.
- Aliyu H, Gorte O, Zhou X, et al. In silico proteomic analysis provides insights into phylogenomics and plant biomass deconstruction potentials of the Tremelalles. Front Bioeng Biotechnol. 2020;8:226.
- Buzzini P, Turchetti B, Yurkov A. Extremophilic yeasts: the toughest yeasts around? Yeast 2018;35(8):487–497.
- Vincent WF. Microbial ecosystems of Antarctica. Cambridge: University Press; 2004.
- Welander U. Microbial degradation of organic pollutants in soil in a cold climate. Soil Sediment Contam Int J. 2005;14(3):281–291.
- Streletskii RA, Kachalkin AV, Glushakova AM, et al. Yeasts producing zeatin. PeerJ. 2019;7:e6474.
- Liu XZ, Wang QM, Theelen B, et al. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud Mycol. 2015;81:1–26.
- Kirk PM, Cannon PE, Minter DW, et al. 2008. Ainsworth & bisby’s dictionary of the fungi. 10th ed. Wallingford: CAB International.
- Takashima M, Nakase T. Dioszegia zsolt emend. In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts, a taxonomic study. Amsterdam: Elsevier; 2001.
- Villarreal P, Carrasco M, Barahona S, et al. Tolerance to ultraviolet radiation of psychrotolerant yeasts and analysis of their carotenoid, mycosporine, and ergosterol content. Curr Microbiol. 2016;72(1):94–101.
- Madhour A, Anke H, Mucci A, et al. Biosynthesis of the xanthophyll plectaniaxanthin as a stress response in the red yeast dioszegia (Tremellales, Heterobasidiomycetes, Fungi). Phytochemistry. 2005;66(22):2617–2626.
- Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73(4):331–371.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322.
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402.
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425.
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press.
- Kimura MA. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791.
- Kurtzman CP, Fell JW, Boekhout T, et al. 2011. The yeast, a taxonomic study. 5th ed. Netherlands: Elsevier. p. 87–110.
- Prillinger H, Lopandic K, Suzuki M, et al. 2011. The yeasts. 5th ed. Netherlands: Elsevier. p. 129–136.
- Fell JW. 2011. The yeasts. 5th ed. Netherlands: Elsevier
- Phaff HJ, Mrak EM, Williams OB. Yeasts isolated from shrimp. Mycologia. 1952;44(4):431–451.
- Crous PW, Wingfield MJ, Chooi YH, et al. Fungal planet description sheets: 1042-1111. Persoonia. 2020;44:301–459.
- Takashima M, Sugita T, Shinoda T, et al. Three new combinations from the Cryptococcus laurentii complex: cryptococcus aureus, Cryptococcus carnescens and Cryptococcus peneaus. Int J Syst Evol Microbiol. 2003;53(Pt 4):1187–1194.
- Chang CF, Huang SY, Lee CF. Vishniacozyma changhuana sp. nov., and Vishniacozyma taiwanica sp. nov., two novel yeast species isolated from mangrove forests in Taiwan. Int J Syst Evol Microbiol. 2019;(3):71.
- Bai FY, Takashima M, Jia JH, et al. Dioszegia zsoltii sp. nov., a new ballistoconidium-forming yeast species with two varieties. J Gen Appl Microbiol. 2002;48(1):17–23.
- Inácio J, Portugal L, Spencer-Martins I, et al. Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Res. 2005;5(12):1167–1183.
- Takashima M, Nakase T. 2011. The yeasts. Netherlands: Elsevier. p. 1747–1757.
