Abstract
The nucleolus is the largest, membrane-less organelle within the nucleus of eukaryotic cell that plays a critical role in rRNA transcription and assembly of ribosomes. Recently, the nucleolus has been shown to be implicated in an array of processes including the formation of signal recognition particles and response to cellular stress. Such diverse functions of nucleolus are mediated by nucleolar proteins. In this study, we characterized a gene coding a putative protein containing a nucleolar localization sequence (NoLS) in the rice blast fungus, Magnaporthe oryzae. Phylogenetic and domain analysis suggested that the protein is orthologous to Rrp8 in Saccharomyces cerevisiae. MoRRP8-GFP (translational fusion of MoRRP8 with green fluorescence protein) co-localizes with a nucleolar marker protein, MoNOP1 fused to red fluorescence protein (RFP), indicating that MoRRP8 is a nucleolar protein. Deletion of the MoRRP8 gene caused a reduction in vegetative growth and impinged largely on asexual sporulation. Although the asexual spores of ΔMorrp8 were morphologically indistinguishable from those of wild-type, they showed delay in germination and reduction in appressorium formation. Our pathogenicity assay revealed that the MoRRP8 is required for full virulence and growth within host plants. Taken together, these results suggest that nucleolar processes mediated by MoRRP8 is pivotal for fungal development and pathogenesis.
1. Introduction
The nucleolus is the largest structure within the nucleus and the site of ribosomal RNA synthesis and nascent ribosome subunit biogenesis in eukaryotic cells [Citation1]. In eukaryotes, three compartments within the nucleolus have been distinguished using electron and light microscopy. Fibrillar center (FC) are surrounded by dense fibrillar component (DFC), which in turn is contained within granular component (GC) [Citation2]. This organization is closely related to the sequential steps of ribosome biogenesis. The FC contain unbound RNA polymerase I, and the DFC contain mostly pre-RNA processing factors. Transcription of rDNA loci occurs at the boundary of the FC and DFC. Transcribed rRNA translocate to the GC region for further maturation and ribosome assembly [Citation3]. Early pre-rRNA precursor are transcribed and cleaved by RNA polymerase I to form mature 28S, 18S and 5.8S rRNAs, which are post-transcriptionally modified through interactions with small nucleolar ribonucleoproteins and many protein-processing factors [Citation4]. Eventually, the processed and modified rRNAs are assembled with several ribosomal proteins and interact with export machinery to be transported to the cytoplasm [Citation5]. In this study, we characterized a gene encoding predicted nucleolar protein, MoRRP8 in Magnaporthe oryzae.
The rice blast fungus, M. oryzae, causes one of the world’s most devastating diseases of cultivated rice. The rice blast disease destroys approximately 10–30% of the rice harvested worldwide each year, posing a serious threat to the global food security [Citation6]. The fungus invades plant cells using a specialized infection cell, called appressorium that forms at the tip of germ tube upon recognition of surface hardness and hydrophobicity [Citation7]. The proliferation following infection leads to the spindle-shape disease lesion, from which asexual spores are produced to re-initiate infection cycles [Citation8].
MoRRP8 is an orthologue of the Rrp8 in Saccharomyces cerevisiae. The ribosome can be divided into two subunits with four rRNA (ribosomal RNA) classes and 70 proteins. The large subunit (LSU) contains 28S, 5.8S, and 5S rRNAs. The small subunit (SSU) has 18S rRNA. The four mature rRNA molecules encode two rRNA genes that are transcribed by two different RNA polymerases. The 18S, 5.8S, and 28S rRNAs are transcribed by RNA polymerase I, called a single transcribed cistron or major transcriptional unit. The 5S rRNA is not normally linked to the ribosomal cistrons and is then transcribed separately by RNA polymerase III [Citation9,Citation10]. ScRRP8 is known as a nucleolar methyltransferase that carry out the m1A base modification at position 645 [Citation11]. ScRRP8 is also involved in A2 cleavage and 40S biogenesis [Citation12]. A yeast strain lacking RRP8 can survive at 30 °C but exhibits strong growth impairment at 19 °C. In these strains, cleavage of pre-rRNA at the A2 site is strongly affected, and cleavage at A0 and A1 sites is slightly inhibited or delayed [Citation13].
MoRRP8 is predicted by NoD web-server and NLStradamus to have nucleolar localization sequences and nuclear localization signal, respectively. The NoD is a web-server that predict nucleolar localization of protein based on the presence of a short stretches of amino acids termed nucleolar localization sequences (NoLS) [Citation14]. The NoD server is the only available prediction tool for detecting the nucleolar localization sequences. The NLStradamus is a prediction tool for nuclear localization signal that are built on simple hidden Markov model [Citation15].
To investigate the roles of MoRRP8 gene in the rice blast fungus, here we first checked the localization of MoRRP8 and then generated gene knock-out mutant of MoRRP8. Using the mutant, we compared the growth, asexual sporulation, conidia germination, appressorium formation, and pathogenicity with those of the wild-type (KJ201) strain.
2. Materials and methods
2.1. Fungal strains and plasmids
The strain used as a wild-type in this study is Magnaporthe oryzae KJ201, which is obtained from the Center for Fungal Genetic Resources (http://cc.aris.re.kr). The plasmids, RBC TA cloning vector and pBC- SK + which is Phagemid vector derived from pBluescript II SK + with a chloramphenicol resistance gene, were used for generating fusion construct.
2.2. Growth condition and media
Total DNA and RNA were isolated from mycelia cultured in liquid complete medium (CM, 0.6% yeast extraction [w/v], 0.6% casamino acid [w/v], 1% sucrose [w/v]). Protoplasts were also prepared from mycelia of cultured in CM. Fluorescence microscopy was carried out using a thin layer of either complete (CM) or minimal medium (MM) placed on a slide glass: CM agar (0.6% yeast extraction [w/v], 0.6% casamino acid [w/v], 1% sucrose [w/v], 1.5% agar powder [w/v]) and MM agar (10% sucrose [w/v], 0.1% Ca(NO3)2·4H2O [w/v], 0.02% KH2PO4 [w/v], 0.025% MgSO4·7H2O [w/v], 0.015% NaCl [w/v], 1.5% agar powder [w/v]). Oatmeal medium agar (OMA, 5% oatmeal [w/v], 2.2% agar powder [w/v]) was used to examine growth, asexual sporulation, conidia germination and appressorium formation. Transformants for gene knock-out and gene complementation were screened by adding hygromycin and geneticin antibiotics to TB3 medium (0.3% yeast extract [w/v], 0.3% casamino acid [w/v], 1% glucose [w/v], 20% sucrose [w/v], 0.8% agar powder [w/v]), respectively.
2.3. Nucleic acid isolation and manipulation
The genomic for PCR-based screening was extracted using the quick method [Citation16]. Total RNA was extracted using the easy-BLUETM Total RNA Extraction Kit. To generate the construct expressing MoRRP8-eGFP fusion protein, T-vector and pBC SK + were used. The native promoter, CDS, and eGFP fragments were amplified and cloned into the T-vector, respectively. Each fragment was retrieved from T-vector using enzyme digestion and ligated into the pBC SK+. Ligation reaction was carried out using EZ-FusionTM HT Cloning kit. Ligated products were cloned into E. coli and resulting colonies were screened for ampicillin resistance and blue-white selection. Colony PCR was performed to identify the true positive colonies. All the primers used in this study were listed in Supplementary Table 1.
2.4. Fluorescence microscopy of the predicted nucleolar localized proteins
MoRRP8-eGFP (MGG_07132-eGFP) were inoculated on complete and minimal medium on the slide in a sealed petri dish containing a towel soaked in distilled water and stored in a 25 °C incubator for seven days. And we observe the fluorescence signal in fungal hyphae under differential interference contrast (DIC) and fluorescence microscopy (Leica DM2500) with GFP (440–520 nm) and RFP filters (515 to 560 nm). The green fluorescence protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet rage. The red fluorescence protein (RFP) is a fluorophore that fluorescence red-orange when excited.
2.5. Generation of the gene knock-out mutants and complemented strains
The 1 kb-upstream and 1 kb-downstream flanking sequences of MoRRP8 was fused with a hygromycin B phosphotransferase (HPH) cassette using a Gibson assembly method (EZ-FusionTM HT Cloning Kit). This construct was introduced into the KJ201 protoplast. As a first screening, hygromycin resistant transformants were picked from TB3 agar medium supplemented with hygromycin B. Then as a secondary screening, genomic DNA of transformants were subject to PCR-based screening. For complementation, MoRRP8-GFP construct and geneticin resistant gene (neomycin phosphotransferase) fragment were introduced into the ΔMorrp8 protoplast, and the resulting transformants were screened by PCR for the presence of MoRR8 gene.
2.6. Multiple sequence alignment and phylogenetic analysis
All nucleotide and amino acid sequences were retrieved from National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov). Sequence alignment and phylogenetic tree construction were done using neighbor-joining method implemented in MEGA7 with 1000 bootstraps [Citation17]. All information related to domain architecture, including the methyltransferase domain was retrieved from NCBI Conserved Domain Search (http://www.ncbi.nlm.nih.gov>structure ≥ cdd ≥ wrpsb).
2.7. Growth, conidiation, and conidiophore development
KJ201 (wild-type), ΔMorrp8, and MoRRP8c were inoculated in oatmeal agar medium and cultured in a 25 °C incubator for nine days. After nine days of growth on the oatmeal agar medium, radial growth of each strain was measured in colony diameter. For comparison of asexual sporulation, hyphae were scraped from the culture plates using distilled water, and then filtered through a miracloth. The number of spores were counted using hemocytometer. For germination and appressorium formation, concentration of spore suspensions was adjusted to 5 × 104 conidial/ml, and 10 µl of spore suspension was placed on a plastic cover glass, which is placed in a sealed petri dish containing wet paper towel and incubated at 25 °C for 2, 4, and 6 h.
2.8. Pathogenicity assay
For pathogenicity assay, spore suspensions were sprayed onto a susceptible rice cultivar Nakdong. The rice seeds were surface sterilized as described by the previous study [Citation18]. The surface-sterilized seeds were allowed to germinate in a sealed petri dish containing wet paper towel. Germinated seeds were planted in soil and grown at 25 °C in plant growth chamber for three weeks. The spore suspensions were adjusted to the 5 × 104 conidial/ml concentration and sprayed onto the rice plants with 250 ppm Tween 80 added to the suspension. The inoculated rice plants were kept in a dew chamber at 25 °C under dark with 100% humidity overnight, and then transferred to the plant growth chamber with 16/8 h light and dark period.
3. Results
3.1. Identification and nucleolar localization of MoRRP8 in M. oryzae
BLASTP search against NCBI nr database using amino acid sequences of MGG_07132 as a query found that the protein encoded by the gene is orthologous to RRP8 in S. cerevisiae (ScRRP8 hereafter). The ScRRP8 is characterized as a rRNA processing protein that is localized to the nucleolus and belongs to class I MTase having Rossmann-like fold with the MTase domain located in the C-terminal part of the protein [Citation19]. Since ScRRP8 acts as a methyltransferase that modifies rRNA, we compared MoRRP8 with other methyltransferases belonging to class I, II, III, IV and V by constructing phylogenetic tree (). This analysis showed that MoRRP8 also belongs to class I methyltransferase, of which all members function as rRNA methyltransferase. The class I to V of AdoMet-dependent families were distinguished by having five structurally different folds [Citation20].
Figure 1. Identification of RRP8 in Magnaporthe oryzae. (A) Phylogenetic analysis of MoRRP8. MoRRP8 was compared phylogenetic genetically to class I, II, III, IV and V methyltransferase. Phylogenetic tree was conducted using neighbor-joining method in Mega7 program. The number above or below branches indicates bootstrap values. Total bootstraps were 1000. Domain architecture of proteins were shown in parallel to the tree; (B) The MoRRP8 tagged eGFP and monitored whether RRP8 localize to the nucleolus using nucleolar marker protein fused to MoNOP1-RFP as a control in M. oryzae. The MoRRP8-eGFP was observed in hyphae under differential interference contrast (DIC) and fluorescence microscopy with RFP, GFP. The first image was observed under 200×, and second image was observed under 400×. The merged panel was generated by merging the GFP and RFP images (scale bar = 10 µm). Information on sequences used in the phylogenetic analysis is as follows: Fusarium oxysporum RRP8 (SCO84821.1); Fusarium keratoplasticum RRP8 (XP_052917695.1); Trichoderma simmonsii RRP8 (QYS93480.1); Schizosaccharomyces pombe RRP8 (CAA93580.1); Saccharomyces cerevisiae RRP8 (NP_010368.5); Colletotrichum siamense SET6 (KAF4836865.1); Schizosaccharomyces pombe CLR4 (NP_595186.1); Saccharomyces cerevisiae DPH5 (NP_013273.1); Saccharomyces cerevisiae SAM4 (NP_015050.1); Saccharomyces cerevisiae HMT1 (NP_009590.1); Sugiyamaella lignohabitans SFM1 (XP_018734810.1); Saccharomyces cerevisiae SFM1 (NP_014664.1); Sugiyamaella lignohabitans EMG1 (XP_018735636.1); Saccharomyces cerevisiae EMG1 (NP_013287.1).
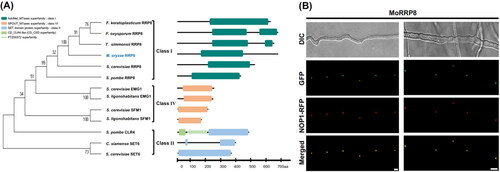
If MoRRP8 is a rRNA processing protein, it is expected to localize in the fungal nucleolus. To check its localization to the nucleolus, we used a nucleolar marker protein MoNOP1 fused to RFP (MoNOP1-RFP) as a control in M. oryzae [Citation21,Citation22]. For comparison, MoRRP8-eGFP was introduced into a strain expressing MoNOP1-RFP. Examination of fluorescence signals under the microscope showed that GFP signals almost completely coincide with RFP signals, indicating that MoRRP8 is a nucleolar protein ().
3.2. Targeted deletion of MoRRP8 gene affects vegetative growth and asexual reproduction of M. oryzae
To understand the contribution of MoRRP8 to fungal development and pathogenesis, we generated deletion mutant of the gene via homologous recombination (Supplementary Figure 1). Compared to the wild-type, the deletion mutant showed significant defect in vegetative growth (). Unlike the wild-type that has grown on average up to 60 mm in diameter, the colony diameter of ΔMorrp8 did not reach 50 mm at nine days post inoculation (). Moreover, the mutant colonies appear to lack pigmentation and go through frequent autolysis (). Introduction of wild-type copy of gene into the mutant was able to restore the radial growth to the wild-type level. However, pigmentation was only partially restored in the complementation strain.
Figure 2. Phenotype of MoRRP8 deletion. (A) Colony morphology, photographs were taken at nine days post inoculation; (B) Colony diameter measurements were taken at nine days post inoculation. The ΔMorrp8 growth is significantly reduced to approximately 11% of the wild type, KJ201 (*, p < .05; **, p < .01; Tukey HSD test n = 3); (C) The conidia count from hyphae grown from individual conidia at nine days. The conidia count of ΔMorrp8 is reduced compared to the wild type, KJ201; (D) The conidiophore formation, these are thin sections after scraping hyphae from the oatmeal agar medium and observed at 100× after 8 h. The conidiophore formation of ΔMorrp8 is reduced compared to the wild type, KJ201.
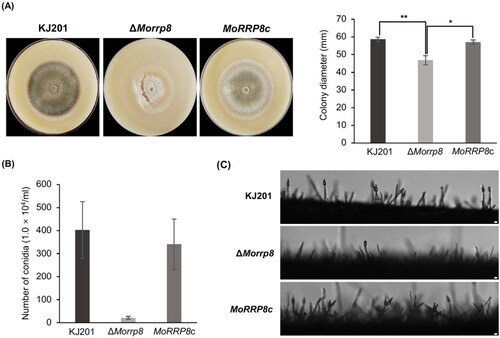
When asexual sporulation was evaluated by quantifying the number of spores produced by each strain, the ΔMorrp8 showed dramatic reduction in asexual sporulation (). Since aerial hyphae mass was reduced in the mutant, we suspected that such drop in sporulation could be attributed to the lack of conidiophores. Close examination of conidiophore formation revealed that indeed conidiophore are rarely found in the mutant ().
3.3. Germination and appressorium formation in the ΔMorrp8
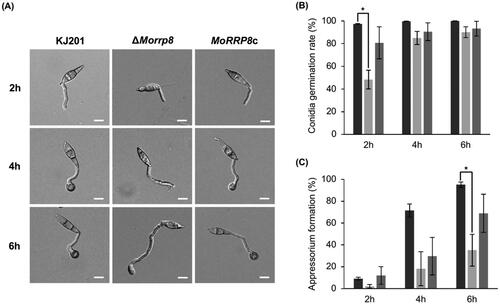
Conidia of the deletion mutant were morphologically indistinguishable from those of wild-type. To see whether the conidia of ΔMorrp8 retain the ability to undergo pathogenic development, we tested conidial germination and appressorium formation. When spore suspensions were placed on plastic coverslip and incubated up to 6 h, the mutant conidia showed delay in germination, compared to the wild-type and complementation strain (). Only about 40% of ΔMorrp8 conidia germinated at 2 h post incubation, although germination rate was comparable between the wild-type and mutant at later hours ().
Figure 3. Effect on MoRRP8 deletion on germination and appressorium formation. (A) the wild-type, KJ201, ΔMorrp8 and MoRRP8c spores at 2, 4 and 6 h under a 200× differential interference contrast (DIC) microscope; (B) at 2 h of germination, ΔMorrp8 was significantly delay Germinating compared to wild-type, KJ201 (*, p < .05; Tukey HSD test n = 3). (C) At 6 h of appressorium formation, ΔMorrp8 was significantly delay forming appressorium compared to wild-type, KJ201 (*, p < .05; Tukey HSD test n = 3).
3.4. Impact of MoRRP8 deletion on fungal pathogenicity
Next, we tested the pathogenicity of the mutant using spray-inoculation method on a susceptible rice cultivar, Nakdong. The wild-type and complementation strains showed numerous lesions at five days post inoculation (dpi), whereas in the mutant, only a few lesions were observed (). As the delay in germination and reduction in appressorium formation cannot fully explain near lack of pathogenicity observed in our assay, we examined if the mutant appressoria are functional using rice sheath assay. This showed that the mutant appressoria are capable of host penetration as evidenced by the presence of penetration pegs and invasive hyphae beneath the appressorium ().
Figure 4. Effect on MoRRP8 deletion on pathogenicity. (A) Lesion development in rice leaves seven days post-spray inoculation; (B) Lesion area measured from post-spray inoculated leaves shown in (A). Graph of the Percentage of lesion area in diseased rice leaves, statistical analysis of diseased lesions after KJ201, ΔMorrp8 and MoRRP8c were all set to the same width (*, p < .05; Tukey HSD test n = 3); (C) Observation of invasive growth using rice sheath assay. Invasive growth in rice sheath cells was monitored under a microscope at 24, 48 h post inoculation (hpi); (D) Lesion development in wound-inoculated rice leaves seven days post-inoculation; (E) Lesion area measured from wound-inoculated leaves shown in (D). Graph showing the length (cm) of lesions formed by KJ201, ΔMorrp8 and MoRRP8c statistics (**, p < .01; Tukey HSD test n = 3).
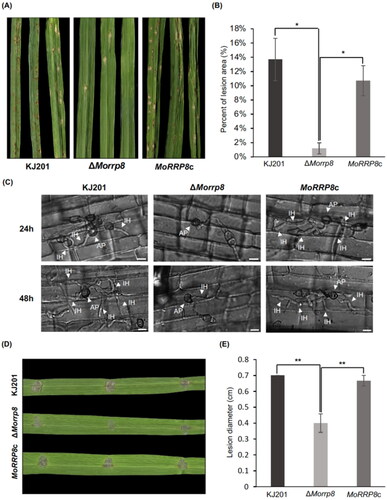
However, unlike the wild-type and complementation strain, growth of invasive hyphae appears to be restricted, suggesting the impairment in invasive growth. Therefore, we further tested if the mutant is able to cause disease symptoms when it is injected into host tissues through wound-inoculation, allowing the fungus to enter the plant cells without appressorium-mediated penetration. Our wound inoculation showed that the mutant develops significantly smaller lesions than the wild-type and complementation strain. Since the decrease in vegetative growth was 10–15%, compared to the wild-type, such reduction in invasive growth (∼ 40% reduction) could not be accounted for by the difference in vegetative growth only (). This result strongly suggests that MoRRP8 plays important roles during invasive growth.
4. Discussion
The nucleolus is an important, membrane-less organelle inside the nucleus for synthesis ribosomal RNA and primary ribosome biogenesis [Citation23]. Such nucleolar functions hinge largely on proteins that are localized to the nucleolus [Citation24]. Much of knowledge about genetic regulation of nucleolar ribosome biogenesis has been learned from the budding yeast and animal cells [Citation25]. Only a few studies have reported the importance of nucleolar proteins in filamentous fungi. For example, Bhabhra et al. demonstrated the requirement of a nucleolar protein, CgrA in integration between morphogenesis and nuclear duplication in Aspergillus fumigatus [Citation26]. More recently, Zhang et al. showed that a serine/threonine kinase CK2 accumulates in nucleoli of M. oryzae as well as in septal pores and appressorium penetration pore, although biological significance of nucleolar accumulation of the protein has not been further investigated [Citation27]. In this study, we investigated a gene encoding a putative nucleolar localized protein, MoRRP8 in the plant pathogenic, filamentous fungi, Magnaporthe oryzae.
Analysis of protein domains showed that MoRRP8 has a methyltransferase domain typically found in class I methyltransferase that are involved in rRNA modification. In addition to the nuclear localization signal (NLS), MoRRP8 is predicted by NoD server to have nucleolar localization sequences (NoLS). Our fluorescence microscopy clearly showed that MoRRP8 is localized to the fungal nucleolus. NoD server was built upon analysis of a set of well-characterized nucleolar proteins in human [Citation28]. Our result, therefore indicates that properties of NoLS are well-conserved features of nucleolar proteins across the different kingdoms in eukaryotes. Sequence similarity and localization strongly suggest that MoRRP8 is a bona fide orthologue of ScRRP8, although its RNA methyltransferase activity has not been demonstrated in our study.
For the deletion mutant of MoRRP8, we found that germination of conidia was delayed. Interestingly, a previous unpublished study using inhibitors showed that germination of spores requires not transcription but translation [Citation29]. In that work, spores were able to germinate when they were treated with transcription inhibitor, while a translation inhibitor, cycloheximide almost completely prevented germination. Based on its orthologous relationship with ScRRP8, MoRRP8-mediated rRNA modification is likely to be required for proper processing of rRNA, which in turn is important for efficient biogenesis of ribosomes in the cell. It is therefore tempting to speculate that lack of MoRRP8 would lead to perturbation in translation, hindering the germination of asexual spores in M. oryzae. In the same vein, such defect in translational process might be able to explain observed reduction in vegetative growth and asexual reproduction.
Other than infection-related development, we found that the mutant is defective in invasive growth as well. To date, roles that nucleolus plays in fungal infection have not been investigated. Our sheath assay showed that the invasive growth of mutant strain is retarded, compared to the wild-type and complementation strains. During vegetative growth, the mutant strain exhibited ∼ 10% reduction in growth rate, and this could not account for the amount of reduction in invasive growth, strongly suggesting the requirement of MoRRP8-mediated process for the fungus to grow inside the host plant. However, how MoRRP8-mediated processes are involved is not clear and warrant more studies in the future. Nevertheless, it is possible that inside the host plant, the fungus demands more energy in order to not only proliferate but also overcome host defense responses for successful colonization. Without proper nucleolar functions, it is reasonable to conjecture that such demands cannot be met by the mutant strain as it is compromised in nucleolar functions.
Supplemental Material
Download JPEG Image (290.4 KB)Supplemental Material
Download MS Word (14.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Pederson T. The nucleolus. Cold Spring Harbor Perspect Biol. 2011;3(3):a000638–a000638. doi: 10.1101/cshperspect.a000638.
- Schwarzacher HG, Wachtler F. The nucleolus. Anat Embryol. 1993;188(6):515–536. doi: 10.1007/BF00187008.
- Raška I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177–235.
- Goodfellow SJ, Zomerdijk JC. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem. 2013;61:211–236.
- Henras AK, Plisson‐Chastang C, O'Donohue MF, et al. An overview of pre‐ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6(2):225–242. doi: 10.1002/wrna.1269.
- Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 2009;27(3):141–150. doi: 10.1016/j.tibtech.2008.12.002.
- Fernandez J, Orth K. Rise of a cereal killer: the biology of Magnaporthe oryzae biotrophic growth. Trends Microbiol. 2018;26(7):582–597. doi: 10.1016/j.tim.2017.12.007.
- Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7(3):185–195. doi: 10.1038/nrmicro2032.
- Mandal RK. The organization and transcription of eukaryotic ribosomal RNA genes. Prog Nucleic Acids Res Mol Bio. 1984;31:115–160.
- Paule MR, White RJ. Survey and summary transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28(6):1283–1298. doi: 10.1093/nar/28.6.1283.
- Sharma S, Hartmann JD, Watzinger P, et al. A single N1-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci Rep. 2018;8(1):11904. doi: 10.1038/s41598-018-30383-z.
- Peifer C, Sharma S, Watzinger P, et al. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 2013;41(2):1151–1163. doi: 10.1093/nar/gks1102.
- Bousquet-Antonelli C, Vanrobays E, Gélugne J-P, et al. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA. 2000;6(6):826–843. doi: 10.1017/s1355838200992288.
- Favre D, Studer E, Michel M. Two nucleolar targeting signals present in the N-terminal part of Semliki Forest virus capsid protein. Arch Virol. 1994;137(1-2):149–155. doi: 10.1007/BF01311181.
- Nguyen Ba AN, Pogoutse A, Provart N, et al. NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinform. 2009;10:1–11.
- Chi MH, Park SY, Lee YH. A quick and safe method for fungal DNA extraction. Plant Pathol J. 2009;25(1):108–111. doi: 10.5423/PPJ.2009.25.1.108.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Bertani I, Abbruscato P, Piffanelli P, et al. Rice bacterial endophytes: isolation of a collection, identification of beneficial strains and microbiome analysis. Environ Microbiol Rep. 2016;8(3):388–398. doi: 10.1111/1758-2229.12403.
- Wlodarski T, Kutner J, Towpik J, et al. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS One. 2011;6(8):e23168. doi: 10.1371/journal.pone.0023168.
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28(6):329–335. doi: 10.1016/S0968-0004(03)00090-2.
- Jansen RP, Hurt EC, Kern H, et al. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol. 1991;113(4):715–729. doi: 10.1083/jcb.113.4.715.
- Tollervey D, Lehtonen H, Carmo‐Fonseca M, et al. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre‐rRNA processing in yeast. EMBO J. 1991;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x.
- Dubois M-L, Boisvert F-M. The nucleolus: structure and function. Funct Nucl. 2016;23:29–49.
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11(3):385–390. doi: 10.1016/S0955-0674(99)80054-4.
- Lin S, Rajan S, Lemberg S, et al. Production of nascent ribosome precursors within the nucleolar microenvironment of Saccharomyces cerevisiae. Genetics. 2022;221(3):iyac070. doi: 10.1093/genetics/iyac070.
- Bhabhra R, Richie DL, Kim HS, et al. Impaired ribosome biogenesis disrupts the integration between morphogenesis and nuclear duplication during the germination of Aspergillus fumigatus. Eukaryot Cell. 2008;7(4):575–583. doi: 10.1128/EC.00412-07.
- 1.Zhang L, Zhang D, Chen Y, et al. Magnaporthe oryzae CK2 accumulates in nuclei, nucleoli, at septal pores and forms a large ring structure in appressoria, and is involved in rice blast pathogenesis. Front Cell Infect Microbiol. 2019;9:113. doi: 10.3389/fcimb.2019.00113.
- Scott MS, Troshin PV, Barton GJ. NoD: a nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinform. 2011;12(1):1–7.
- Zeigler RS, Leong SA, Teng PS, editors. Rice blast disease. Wallingford: CABI; 1994.
