Abstract
Fungi are cosmopolitan and they occupy diverse niches as consumers, producers, and decomposers. They play critical roles in the environment by enabling nutrient cycling and generating a plethora of secondary metabolites. This study aimed to identify and characterize fungal strains isolated from diverse sources on Muui Island, Republic of Korea. In 2023, a total of 86 fungal strains were collected and examined. Investigation of the morphological features and phylogenetic analyses of multiple barcode loci identified one putative novel species and two species previously unrecorded in the Republic of Korea: Colletotrichum sp., Colletotrichum guizhouense, and Fusarium brachygibbosum. This study provides a comprehensive description of their molecular phylogenies and morphological characteristics. These findings will contribute to the existing knowledge about fungal species in the Republic of Korea and future research on the fungal diversity on Muui Island.
1. Introduction
Fungi are crucial to our ecosystem, serving as consumers, producers, and decomposers. Fungi have a profound impact on our daily lives, including their utilization in agriculture, food, and the pharmaceutical field. Exploration of fungal resources through collecting and culturing fungi from the environment provides valuable insights into fungal taxonomy, environmental roles, and industrial applications. Consequently, the discovery of new or previously unrecorded species continues at the national level as well as global level. In the Republic of Korea, a total of 4927 official fungal species have been discovered and registered [Citation1].
Muui Island is an island located in Incheon, Republic of Korea, covering an area of 9.4 km2 with a total coastline length of 31.6 km. The topography of Muui Island consists mainly of mountainous terrain, with three mountains (Dongsan, Guksa Peak, and Horyonggok) and two beaches (Hanae Beach and Silmi Beach) along with tidal flat terrain. In addition, Korean hornbeam (Carpinus turczaninovii), which is mainly distributed in Southeast Asia with high biodiversity, forms forest stands in this area. Muui Island harbors a rich variety of ecosystems, coexisting with the mountains, beaches, and tidal flats, and previous studies have investigated the distribution of native plant species on the island [Citation2]. However, research focusing on indigenous fungi remains lacking.
Thus, this study aimed to identify and characterize the morphological and taxonomic features of fungal species in Muui Island, with the objective of understanding the fungal communities in the area and finding undescribed or unrecorded fungal species. A total of 86 fungal specimens were collected from Muui Island in April 2023. The fungal communities were composed of 21 genera and 46 putative species. Through phylogenetic analyses of multiple DNA barcode loci one putative novel species and two species previously unrecorded in the Republic of Korea were newly identified. This study provides detailed descriptions of the morphological observations and molecular phylogenetic analyses to report them as newly recorded species in the Republic of Korea.
2. Materials and methods
2.1. Sampling and fungal isolation
In April 2023, 31 samples from various sources, including plants and soil, were collected from three sites in Muui Island (located in front of Incheon) (, ). The collected samples were cut into 10 mm pieces and surface sterilized using one of two methods; either soaking in 1% sodium hypochlorite for 1 min, followed by rinsing with sterile water three times, or rinsing with sterile water three times only. Five or six sterilized pieces were placed on potato dextrose agar (PDA; Difco, Sparks, MD, USA) plates (90 mm diameter) containing 100 µg mL−1 of ampicillin and 100 µg mL−1 of streptomycin, and the plates were incubated at 25 °C for 3–5 days. To isolate fungal strains from soil samples, 10 g of soil was diluted in 50 ml of sterile distilled water and agitated in a rotary shaker at 150 rpm for 30 min. Subsequently, 100 µL of supernatant was spread onto PDA plates containing 100 µg mL−1 of ampicillin and 100 µg mL−1 of streptomycin and incubated at 25 °C for 3–5 days. Fungal colonies were transferred to fresh PDA plates. All isolates were pure-cultured by hyphal-tip transfer and stored in 20% glycerol at −80 °C until use. Representative isolates annotated as unrecorded species in the Republic of Korea, listed in , were deposited at the Honam National Institute of Biological Resources of the Ministry of Environment in Korea (HNIBR) under deposit numbers HNIBRFG4468, HNIBRFG4472, and HNIBRFG4478.
Figure 1. Representative photographs illustrating examples of the sampling sites in this study. (A) Geographical location map of the sampling sites in Muui Island, Republic Of Korea; images of the collected samples at each sampling site were represented as (B) site 1; (C) site 2; and (D) site 3; respectively.
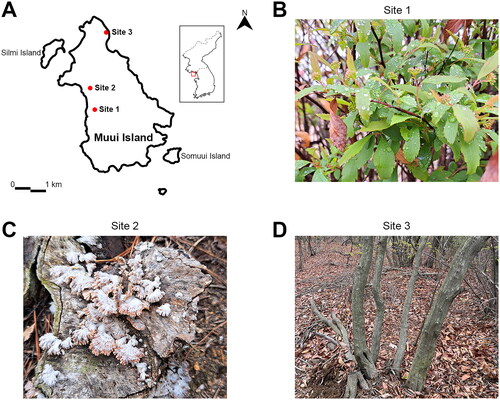
Table 1. Three sampling sites and 31 sampling substrates.
Table 2. Putative novel species and previously unrecorded species with details of HNIBR deposit number of the strains and GenBank accession numbers of the sequences.
2.2. Molecular identification and phylogenetic analyses
Fungal genomic DNA was extracted following the standard protocol [Citation3]. Initially, the internal transcribed spacer (ITS) region of 18S-28S nuclear ribosomal DNA (nrDNA) was amplified using the primer pair ITS4/ITS5 [Citation4]. Polymerase chain reaction (PCR) was performed in 50 µL reactions with Taq DNA polymerase (Takara Shuzo, Japan), 20 pmol of each primer, and approximately 50 ng of template DNA, as previously described [Citation4]. Amplified PCR products were purified using the α+ SolutionTM GEL/PCR Purification Kit (Alphagen, Changzhi, Taiwan) and subsequently sequenced (Macrogen, Seoul, Korea).
DNA sequences were analyzed using SeqMan Pro (DNAStar, Madison, WI, USA) and searched using nBLAST in the NCBI database (https://blast.ncbi.nlm.nih.gov). Species identification was assigned to those with over 97% similarity in BLAST using the NCBI database. For accurate species identification, additional fungal barcode regions for each species were identified using reference sequences from GenBank. The translation elongation factor-1α (tef1) sequences were amplified using the primer set EF1-728F/EF-2 for Colletotrichum sp. and Fusarium brachygibbosum [Citation5]. The β-tubulin (tub2) sequences of Colletotrichum guizhouense and Colletotrichum paraendophytum were amplified using the primer set T1/Bt2b [Citation6,Citation7]. Manganese-superoxide dismutase (sod2) and chitin synthase (chs-1) sequences of C. guizhouense were amplified using the primer sets SOD625F/SOD625R and CHS-79F/CHS-354R, respectively [Citation5, Citation8]. The sequences of the unrecorded species were deposited in GenBank and listed in . The forward and reverse sequences generated from Sanger sequencing were aligned and then trimmed using MEGA_X [Citation9]. Maximum likelihood analyses were performed with the general time-reversible model GTR + GAMMA + I with 1000 bootstrap replicates using RAxML [Citation10].
2.3. Morphological observation
Radial growth and colony morphology of three unrecorded species were examined on PDA plates six days after inoculation. Additional media, including synthetic nutrient-poor agar (SNA) and oatmeal agar (OA) were used for further morphological observation. Spores were collected from cultures grown on PDA for 14 days. The structure of spores was observed under a light microscope at 400× magnification (Leica DM500, Jena, Germany). The average dimensions of spores were calculated based on measurements of 30 spores per isolate using ImageJ [Citation11].
3. Results
3.1. Survey of fungal distributions in Muui Island
To investigate the fungal diversity of Muui Island, 31 samples from various sources across three sites were collected in April 2023 (, ). Especially, Site 3 harbors forest stands of Korean hornbeam, a native species of Korea ((D)). We successfully isolated and identified a total of 86 fungal strains, which represented 46 putative species within 21 genera with high sequence similarity to the ITS sequences of reference strains, (Supplementary Table S1). The most frequently isolated genus was Fusarium (14 isolates) followed by Pestalotiopsis (13 isolates). Notably, three isolates were recognized as potential new species that have not been previously recorded in Korea.
3.2. Species identification
Multiple sequence alignment was performed using DNA barcode regions ITS, tub2, tef1, chs-1, or sod2 to identify the species of three isolates that are considered unrecorded in Korea (, ). Through maximum likelihood (ML) methods, three isolates were identified into three species: C. guizhouense (27-B-4) ((A)), Colletotrichum sp. (12-B-2) ((B)), and Fusarium brachygibbosum (1-B-1) ((C)).
Figure 2. Phylogenetic trees based on Maximum likelihood (ML) analysis of a combined DNA dataset of barcode gene sequences for the strain 27-B-4, 12-B-2, and 1-B-1 in relation to closely related taxa. (A) Phylogenetic analysis of Colletotrichum isolates in the spaethinum complex based on a ML tree of the combined the ITS and tub2 sequence. (B) Phylogenetic analysis of Colletotrichum isolates in the graminicola complex based on a ML tree of the combined the ITS, tub2, chs-1, and sod2 sequence. (C) Phylogenetic analysis of Fusarium isolates based on a ML tree of the combined the ITS and tef1 sequence. Bootstrap values over 70 are presented at the nodes. The scale bar represents the number of nucleotide substitutions per site. The species newly discovered in this study were highlighted in bold.
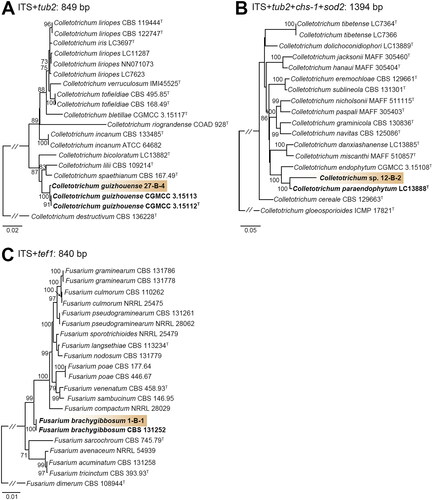
Phylogenetic analysis revealed that 27-B-4 classified in Colletotrichum, formed a monophyletic group with C. guizhouense CGMCC 3.15112 and CGMCC 3.15113 (sequence similarity for ITS = 100%, tub2 = 99.5%; bootstrap support = 100%). 12-B-2 formed a distinct clade in the C. graminicola species complex and showed low similarity with the closest strain C. paraendophytum (LC13888) (sequence similarity for ITS = 87.1%, tub2 = 95.9%, chs-1 = 99.4%, and sod2 = 97.9%; bootstrap support = 100%). Furthermore, ML analysis using each barcode gene sequence revealed that its taxonomic position is distinct from other identified species of Colletotrichum (Supplementary Figure S1). 1-B-1 formed a clade with the type strain (CBS 131252) of Fusarium brachygibbosum (sequence similarity for ITS = 100% and tef1 = 99.8%; bootstrap support = 99%).
3.3. Taxonomy
Colletotrichum guizhouense G. Tao, Zuo Y. Liu and L. Cai (2013) ( and ).
Figure 3. Morphological characteristics of the three fungal species in this study. (A) Colony morphology of Colletotrichum spp. after 6 days at 25 °C, from left to right potato dextrose agar (PDA), oatmeal agar (OA), synthetic nutrition-poor agar (SNA), and microscopic images of conidiophore and conidia. Scale bar: 20 µm. (B) Colony morphology of Fusarium brachygibbosum after 6 days at 25 °C, from left to right PDA, OA, SNA, and microscopic images of phialide and conidia. Scale bar: 20 µm.
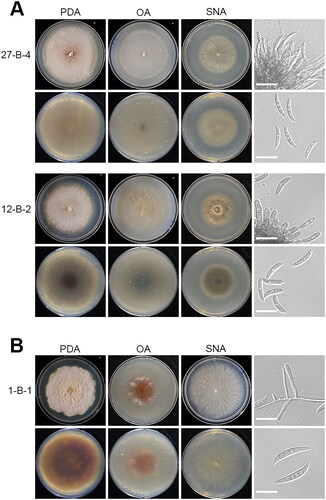
Description: Colonies on PDA 75–78 mm diam in 6 d, flat with lobate edge, brown with white edge, texture cottony, reverse white, exudates absent, soluble pigments absent. Sporulation low, conidia light brown. On OA 62–66 mm diam in 6 d, flat with feathery edge, pale brown to white, reverse white, texture cottony, exudates absent, soluble pigments absent. Sporulation low, conidia light brown. On SNA 52–56 mm diam in 6 d, flat with lobate edge, white, texture cottony, reverse white, exudates absent, soluble pigments absent. Vegetative hyphae septate, branched, straight or slightly bent, hyaline to light grey. Asexual morph formed on PDA and OA. Conidiophores septate, usually branched, cylindrical, hyaline to pale brown. Conidia aseptate, single celled, fusiform, hyaline; conidial length 19.2 to 27.0 µm (mean ± SD = 23.6 ± 1.9 µm, n = 30). Appressoria not observed.
Strain examined: 27-B-4 isolated from grasses at site 3 in Muui Island, Republic of Korea.
Notes: 27-B-4 is morphologically similar to the type strains CGMCC 3.15112 and CGMCC 3.15113 of Colletotrichum guizhouense, except for the following features [Citation12]. 27-B-4 has darker mycelia in the center on PDA and grows faster on PDA (75-78 mm on 6 d vs 56-74 mm on 7 d) [Citation12].
Colletotrichum sp. ( and ).
Description: Colonies on PDA 71–78 mm diam in 6 d, flat with lobate edge, white, texture fluffy, reverse white to slightly gray, exudates absent, soluble pigments absent. Sporulation low, conidia light grey. On OA 65–68 mm diam in 6 d, flat with feathery edge, greenish grey, texture cottony, reverse greenish grey. Sporulation low, conidia light grey. On SNA 46-48 mm diam in 6 d, flat with erose edge, greenish brown with white edge, texture cottony, reverse greenish brown with white edge. exudates absent, soluble pigments absent. Vegetative hyphae septate, branched, hyaline to light brown. Asexual morph formed on PDA and OA. Conidiophores septate, usually branched, cylindrical, hyaline to light brown. Conidia aseptate, slightly curved, hyaline; and conidial length 19.0–24.5 µm (mean ± SD = 21.5 ± 1.6 µm, n = 30). Appressoria not observed.
Strain examined: 12-B-2 isolated from grasses at site 2 in Muui Island, Republic of Korea.
Notes: 12-B-2 shows similar colony and conidial morphology to the type strain LC13888 of Colletotrichum paraendophytum [Citation13]. However, 12-B-2 is phylogenetically distinguished from the type strain LC13888 of C. paraendophytum and has brighter mycelia than the type strain on PDA. Interestingly, 12-B-2 showed low sequence similarity with ITS (87.1% similarity), tub2 (95.9% similarity) with the closest type strain LC13888 of C. paraendophytum. Acquiring more strains is essential to define 12-B-2 as a novel specimen for accurate identification in further study.
Fusarium brachygibbosum Padwick (1945) ( and ).
Description: Colonies on PDA 70–75 mm diam in 6 d, flat with erose edge, yellowish white, texture fluffy, reverse reddish brown, exudates absent, soluble pigments absent. On OA 48–55 mm diam in 6 d, flat with entire to undulate edge, red with white edge, texture cottony, reverse red to white edge, exudates absent, soluble pigments absent. Sporulation moderate, conidia white. On SNA 74-78 mm diam in 6 d, flat with undulate to lobate edge, texture fluffy, reverse white, exudates absent, soluble pigments absent. Sporulation moderate, conidia white. Vegetative hyphae septate, unbranched or irregularly branched bearing terminal phialide, hyaline to light yellow. Asexual morph formed on OA and SNA. Macroconidia slightly curved toward the basal part, three to five septa, hyaline and conidial length 22.1–48.3 µm (mean ± SD = 36.3 ± 7.3 µm, n = 30).
Strain examined: 1-B-1 isolated from plant roots at site 1 in Muui Island, Republic of Korea.
Note: The overall morphological characteristics of 1-B-1 are similar to the previously reported strain of Fusarium brachygibbosum [Citation14,Citation15]. However, the length of macroconidia of 1-B-1 was larger than that of the strain of F. brachygibbosum (22.1–48.3 µm vs. 15.2–22.0 µm) [Citation14].
4. Discussion
In this study, we investigated the fungal diversity on Muui Island and discovered three species previously unrecorded in Korea. A total of 86 fungal strains were putatively identified through nBLAST using ITS sequences, and accurate identification of three unrecorded species was achieved by constructing phylogenetic trees with multiple barcode loci. We selected the DNA barcode regions typically used for each fungal genus in this study. The tef1 gene was proposed as specific marker for Fusarium [Citation16], and the tub2 gene for Colletotrichum spp. [Citation13]. The three species that were previously unrecorded in Korea were found to belong to two different orders; Glomerellales and Hypocreales. We also found strains of Mucor variicolumellatus and Sistotrema brinkmannii that had been described before [Citation17,Citation18] but were not present in the NIBR database (Supplementary Figure S2).
Since the fungal species were primarily isolated from or found near plants, we explored the possibility that the unreported species may be plant pathogens. Fusarium brachygibbosum is reported as a dieback pathogen of Euphorbia larica and a pathogen of potato tubers [Citation19,Citation20]. Therefore, it is plausible that they may act as plant pathogens in the isolation sites. However, the direct association between the isolated fungal species and their source of origin is yet to be confirmed. Therefore, further studies are needed to validate their pathogen-host interaction and pathogenicity. In conclusion, our study uncovers three unrecorded fungal species on Muui Island, significantly contributing to our understanding of fungal diversity and distribution in Korea.
Revised_Supplemental material_Clean.docx
Download MS Word (1 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- National Institute of Biological Resources. National list of species of Korea, 2022. [cited 2023 July 25]. Available from: https://kbr.go.kr/.
- Kim H-J, Son DC, Lee D-H, et al. Flora of vascular plants in Mueuido (Incheon), Korea. Korean J Environ Biol. 2016;34(4):246–256. doi: 10.11626/KJEB.2016.34.4.246.
- Leslie JF, et al. The Fusarium laboratory manual. Ames, Iowa: Blackwell Publishing. 2006. p. 57–77.
- White TJ, Bruns TD, Lee SB, et al. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press. 1990. p. 315–322.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999;91(3):553–556. doi: 10.2307/3761358.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995.
- O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116. doi: 10.1006/mpev.1996.0376.
- Crouch JA, Clarke BB, Hillman BI. Unraveling evolutionary relationships among the divergent lineages of Colletotrichum causing anthracnose disease in turfgrass and corn. Phytopathology. 2006;96(1):46–60. doi: 10.1094/PHYTO-96-0046.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Edler D, Klein J, Antonelli A, et al. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol. 2021;12(2):373–377. doi: 10.1111/2041-210X.13512.
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089.
- Tao G, Liu Z-Y, Liu F, et al. Endophytic Colletotrichum species from bletilla ochracea (Orchidaceae), with descriptions of seven new speices. Fungal Divers. 2013;61(1):139–164. doi: 10.1007/s13225-013-0254-5.
- Liu F, Ma ZY, Hou LW, et al. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud Mycol. 2022;101(1):1–56. doi: 10.3114/sim.2022.101.01.
- Jayawardena R.S, Hyde KD, Wang S, et al., Fungal diversity notes 1512–1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers, 2022. 117(1): p. 1–272. doi: 10.1007/s13225-022-00513-0.
- EFSA Panel on Plant Health (PLH). Pest categorisation of Fusarium brachygibbosum. Efsa J. 2021;19(11):e06887.
- Boutigny A-L, Gautier A, Basler R, et al. Metabarcoding targeting the EF1 alpha region to assess Fusarium diversity on cereals. PLoS One. 2019;14(1):e0207988. doi: 10.1371/journal.pone.0207988.
- Paul NC, Park S, Liu H, et al. Fungi associated with postharvest diseases of sweet potato storage roots and in vitro antagonistic assay of Trichoderma harzianum against the diseases. JoF. 2021;7(11):927. doi: 10.3390/jof7110927.
- Kim D-H, Kim S-H, Kwon S-W, et al. The mycobiota of air inside and outside the meju fermentation room and the origin of meju fungi. Mycobiology. 2015;43(3):258–265. doi: 10.5941/MYCO.2015.43.3.258.
- Al-Mahmooli IH, Al-Bahri YS, Al-Sadi AM, et al. First report of euphorbia larica dieback caused by Fusarium brachygibbosum in Oman. Plant Dis. 2013;97(5):687–687. doi: 10.1094/PDIS-09-12-0828-PDN.
- Jasarevic M, Catalani A, Morales-Rodríguez C, et al. First report of Fusarium brachygibbosum causing rots on potato tubers in Italy. J Plant Pathol. 2022;104(4):1591–1591. doi: 10.1007/s42161-022-01221-z.
