Abstract
The regulation of the majority of cold-regulated genes in plants is mediated by CBF (C-repeat binding factors) transcription factor family. Natural differences in frost tolerance (FT) of wheat have been mapped to the Fr-2 (Frost Resistance-2) locus on chromosome group 5 and are associated with variation in threshold induction temperatures and/or transcript levels of CBF genes.
Abstract
This study used real-time reverse-transcription polymerase chain reaction (qRT-PCR) to compare the relative expression levels of four T. aestivum CBF genes (TaCBF15.2, TaCBFA19, TaCBFA2 and TaCBFD21) in crown tissue of two Bulgarian hexaploid winter wheat cultivars (Milena and Russalka) with distinct levels of low-temperature (LT) tolerance but same vernalization requirement, and the spring cultivar Chinese Spring.
Abstract
The transcription profiles of the selected TaCBF genes showed that they are induced by cold treatment at 2 °C. Analysis of transcript abundance revealed that the four TaCBF genes were expressed at higher levels in the frost tolerant Milena than in the susceptible Russalka. Largest differences (fivefold and fourfold) in expression levels between both winter cultivars were observed in two of the analysed genes, TaCBF15.2 and TaCBFA19, respectively. The higher steady-state expression levels of TaCBF genes before the onset of the LT treatment in Milena, combined with stronger induction by cold treatment, suggest that these molecular responses to LT are associated with superior FT development capacity.
Abstract
The results expand our understanding of the molecular mechanisms underlying LT acclimation in Bulgarian wheat and can be used for development of functional markers for improvement of FT wheat-breeding programmes.
Introduction
Low temperature (LT) is one of the major abiotic stress factors that limit the growth, productivity and geographical distribution of agricultural crops,[Citation1] including temperate cereals. Even in the established agricultural production areas, seasonal or episodic freezing events can lead to significant losses in yield and quality of wheat. Winter wheat with higher yield and quality potential has to withstand freezing temperatures during the winter. Currently, due to the increased necessity of feeding an ever-expanding human population, one of the main breeding efforts worldwide is the development of wheat cultivars with better adaptation to cold. Greater LT tolerance allows more flexible farm management, environmentally friendly production systems, lower herbicide costs and energy requirements and, finally, increased crop moisture utilization and productivity.[Citation1] Identifying and understanding the genetic and molecular mechanisms of LT adaptation are crucial for improvement of winter wheat survival and thereby the production. Once the LT tolerance pathways and the roles of important cold tolerance genes are elucidated, directed breeding strategies can be developed to increase the LT tolerance in winter wheat.
Autumn-sown winter cereals acquire tolerance to freezing temperatures and become vernalized through exposure to low non-freezing temperatures. The level of accumulated LT tolerance depends on the cold acclimation rate and factors controlling the time of floral transition at the shoot apical meristem.[Citation2] Cold acclimation and vernalization are indispensable for winter wheat cultivars to both survive freezing temperatures in the winter and switch from vegetative to reproductive stage in the spring. During cold acclimation, winter cereals undergo large-scale changes in their transcriptome, which in turn activates the production of an array of proteins to aid the plant's survival during subsequent freezing stress.[Citation3,4]
The vernalization delays flowering until the end of winter and protects the sensitive floral primordia.[Citation5] In wheat this process is regulated by a genetic pathway that involves at least three genes [TaVRT-1 (VRN-1), VRN-2, TaVRT-2, TaFT1(VRN3)]. The vernalization locus VRN-1 has been shown to be involved in the LT requirement, and the Vrn-A1 gene localized on chromosome 5A has been suggested to be a stronger spring/winter habit determinant [Citation6] than the homoelogous Vrn-B1 (5B) and Vrn-D1 (5D).[Citation7] Association between the vrn-1 winter allele and greater freezing tolerance and winter hardiness has been documented.[Citation8,9] Molecular genetic studies have shown very tight genetic linkage between the VRN1 loci and a major quantitative trait locus (QTL), the frost tolerance 1 (Fr-1) locus, associated with resistance to cold stress.[Citation10–12] Fr-1 has a key role in the freezing tolerance of vegetative tissues. Because the Fr-1 allele conferring greater freezing tolerance and winter hardiness co-segregates with the vrn-1 winter allele, it has not been well clarified whether this locus is just a pleiotropic effect of VRN-1 or it is a separate gene.[Citation13,14] Later results indicate that the Vrn–Fr-1 region consists of two distinct, but closely linked loci that control both frost tolerance and vernalization.[Citation15] However, despite the progress in the mapping of the Vrn1–Fr1 intervals in common wheat, the isolation of the key Fr-1 gene on this chromosomal region has yet to be done. A second distinct locus, Fr-2, affecting freezing tolerance has been detected in a number of wheat and barley mapping populations.[Citation16–19] This locus is located about 30 cM proximal to locus VRN-1 on the long arm of chromosome 5.[Citation16–19] Fr-2 is coincident with a cluster of genes encoding C-repeat binding factors (CBFs). The CBFs are transcriptional activator proteins that regulate pathways affecting cold acclimation and freezing tolerance.[Citation20–23] They belong to the AP2/EREBP transcription factor family and induce COR (cold-regulated) gene transcription by binding to C-repeat/dehydration-responsive elements (CRT/DRE) in the regulatory regions of the COR genes.[Citation24] CBFs have been implicated in the transcriptional control of many genes, including those involved in phosphoinositide metabolism, biosynthesis of osmolytes, Reactive Oxygen Species (ROS) detoxification, membrane transport, hormone metabolism and signalling.[Citation4,Citation22,Citation25,26]
In addition to their induction by various abiotic stimuli, the expression of CBFs is affected by photoperiod [Citation27] and diurnal cycles.[Citation28,29] Some CBF genes are expressed at low levels in non-stressed conditions which may lead to continual, albeit low, accumulation of their target genes.[Citation5,Citation15,Citation27,28,Citation30–32] Over-expression of the wheat CBF transcription factors increased the freezing tolerance of transgenic plants.[Citation33–35] The relatively large CBF families identified in most cereals were divided into four major phylogenetic groups, consisting of two or more sub-groups each.[Citation28] The CBF subgroups IIIc, IIId, IVa, IVb, IVc and IVd appear to be unique in Pooideae.[Citation28,Citation36,Citation37] Among them, five were suggested to be involved in the development of higher LT tolerance in Triticeae crops with a winter growth habit.[Citation28] Several CBF genes have been characterized, including 17 from barley,[Citation36] 15 from T. monococcum,[Citation37] 37 from hexaploid (6x) wheat (T. aestivum) [Citation28] and 11 from rye (S. cereale).[Citation29] More recent studies [Citation38] revealed more complex organization of the CBF family in 6x wheat, which consists of at least 65 CBF genes, 27 of which are paralogs with 1–3 homeologous copies for the A, B and D genomes. Sixty of them were shown to be expressed in the frost tolerant cultivar Norstar. At least 11 different CBF genes have been mapped within a small 0.7–0.8 cM region encompassing the Fr-2 locus in both T. monococcum and barley.[Citation18,Citation37,Citation39,40] Subsequently, high-density mapping of this region identified three CBF genes (CBF12, CBF14 and CBF15) most probably responsible for the observed differences in cold tolerance in T. monococcum.[Citation41] Although there are indications that the CBF genes cluster on the homeologous group 5 chromosomes of Triticeae tribe, and coincide with the QTL for LT tolerance,[Citation16,17,Citation19,Citation29,Citation37,Citation39] it is still not clear which CBF genes have a stronger effect on LT tolerance in 6x winter wheat and whether their effect is additive.
In Bulgaria, due to specific agro-climatic conditions, breeding for cold tolerance in 6x wheat has a long tradition. A large number of freezing tolerant cultivars were developed during the last 20 years. However, the genetic mechanism underlying the cold tolerance in Bulgarian wheat is still unknown. Unravelling of the mechanism of natural differences in frost tolerance is an important issue for further improvement of breeding strategies for this trait.
The objective of this study was to identify TaCBF genes with major effect on the frost tolerance (FT) of Bulgarian hexaploid winter wheat cultivars. These can prove useful as functional markers in marker-assisted selection for increased frost tolerance.
Materials and methods
Plant material
Two wheat (Triticum aestivum L.) cultivars, Milena and Russalka, from the gene bank of the Dobrudzha Agriculture Institute (General Toshevo, Bulgaria) as well as Chinese Spring (CS) were used in this study. The winter habit (vrn-A1/vrn-A1, vrn-B1/vrn-B1, vrn-D1/vrn-D1) of Bulgarian cultivars and the spring habit of CS (Vrn-D1/Vrn-D1) were verified by polymerase chain reaction (PCR) in previous studies [Citation42–44] using the perfect molecular markers developed by Yan et al. [Citation45] and Fu et al.[Citation46] Both Bulgarian cultivars have the same vernalization requirements of 40 days. The photoperiod insensitive phenotypes (Ppd-D1a) of both winter cultivars were confirmed by PCR with the allele-specific primers described by Beales et al.[Citation47]
Frost tolerance test
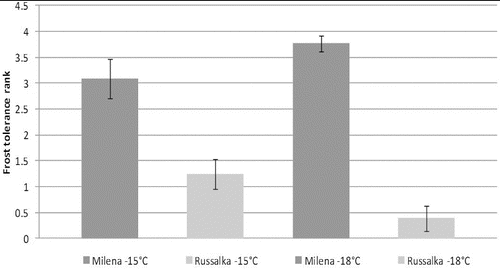
Screening for frost tolerance of Bulgarian cultivars was performed at the Agricultural Institute, Centre for Agricultural Research, Hungarian Academy of Sciences, Martonvásár (Hungary), following a laboratory method described by Vágújfalvi et al.[Citation19] Seeds were germinated in Petri dishes to achieve the desired size of the seedlings (1.5 cm coleoptile and 1.5 cm roots). They were plotted in wooden boxes in a randomized block design arrangement and were grown in a growth chamber at 15 °C/10 °C (day/night), 75% relative humidity and 260 μmolm−2 s−1 light intensity. The hardening started when the temperature was reduced to 10 °C/5°C for two weeks, then to 5 °C/0 °C for another two weeks and to +2 °C/−2 °C for one week. Then the plants were gradually subjected to 24 h freezing at two different temperatures, −15 °C and −18 °C. After freezing, the temperature was gradually increased to 17 °C/16 °C and the leaves were cut several centimetres above the soil. Frost tolerance was estimated as the assessment of the re-growth of the plants scored on a scale running from 0 (dead) to 5 (undamaged, tillering plants).
Plant growth conditions and cDNA synthesis
Seeds of the winter cultivars Milena and Russalka and the spring cultivar CS were germinated on wet filter paper in Petri dishes at 20 °C for 1 day, subsequently at 4 °C for 2 days to synchronize the germination and then moved back at 20 °C for additional 2 days. The seedlings were transferred to hydroponics trays containing modified Hoagland solution [Citation48] for 8 days at 22 °C/16 °C under a 16 h day/8 h night photoperiod, where the illumination stared at 8:00 with a light intensity of 250 μmol m−2 s−1 and at 73% of humidity in a growth chamber. At the end of this period 1–3 g of the crown tissue (less than 1 cm long sections at the base of the crown) were sampled and individually frozen.
Cold treatment (2 °C) was initiated by changing the temperature in the growth chamber at 8:00 am (with lights on as described above), and maintained for 7 h, followed by transfer to 22 °C for additional 24 h of de-acclimation. Control and cold treated plants were sampled at multiple time points during cold treatment as shown in . The rapidly harvested crown tissue was immediately frozen in liquid nitrogen and stored at −70 °C until use for RNA extraction.
Table 1. Sampling scheme of crown tissue.
RNA was extracted from approximately 100 mg leaf tissue, using Trizol (Invitrogen™) following the manufacturer's instructions. The concentration and quality of RNA were assessed by NanoDrop 2000 (Thermo Scientific) and by gel electrophoresis, respectively. The RNA was treated with TURBO DNase (Ambion®) to remove possible DNA contamination in RNA samples. The absence of genomic DNA was confirmed by gel electrophoresis and control real-time quantitative reverse-transcription PCR (qRT-PCR) reactions lacking reverse transcriptase enzyme. First strand cDNA synthesis was performed on 5 μg total RNA, using RevertAid H Minus M-MuLV reverse transcriptase (Fermentas, 50 U), Ribonuclease Inhibitor (Fermentas, 40 U), oligo (dT)18 primers (1 μg) in 20 μL. First strand cDNA samples were diluted in water to a concentration of 40 ng/μL and stored at −70 °C until use.
Real-time qRT-PCR
The transcript accumulation of each gene was detected by qRT-PCR using TaqMan fluorescence detection on a 7300 Real-Time PCR System (Applied Biosystems®). The gene-specific primers and the fluorescent TaqMan probes were designed by Primer Express® Software Version 2.0 (Applied Biosystems®) using the nucleotide sequences of the selected CBF genes (EF028764, EF028766, EF028769 and EF028775) [Citation28] (). Both gene-specific primers and the dual labelled TaqMan probes (FAM and TAMRA) were purchased from Applied Biosystems®.
Table 2. Primers used in qRT-PCR.
PCR mix was performed in a total volume of 25 μL containing 120 ng single-stranded cDNA as a template, 12.5 μL 2xTaqMan Universal PCR Master mix (Applied Biosystems®), 0.9 pmol/μL of each forward and reverse primer and TaqMan probe (0.2 pmol/μL). The cycling conditions were as follows: 10 min at 95 °C, followed by 50 cycles of 95 °C for 15 s/60 °C for 60 s. All PCR reactions were repeated twice.
The constitutively expressed gene α-actin (CJ902800) [Citation49] was used as an internal control. Raw Ct values (difference in the number of PCR cycles required to reach the log phase of amplification) from two repetitions were imported into BioRad Gene Expression Macro Version 1.1 software for expression analysis. The relative expression level was determined by using the 2−ΔΔCt method,[Citation50] where the Ct values of the target gene, normalized to actin (ΔCt), were expressed relative to the Ct values of the ‘calibrator’ CS at 0 h.
Results and discussion
Frost tolerance rank of Bulgarian winter wheat cultivars
The frost tolerance test of the selected cultivars with winter growth habit (vrn-1, vrn-3) was performed in Hungary (2008) under controlled conditions, using two freezing temperatures (−15 ºC and −18 ºC). According to the results from the frost test, Milena proved to be highly frost tolerant, while Russalka was identified as frost sensitive (). These results were confirmed by Tsenov et al. [Citation51] and Ganeva et al. [Citation52] through a multi-year frost tolerance test using the field-laboratory method [Citation53] at the Dobrudzha Agricultural Institute, G. Toshevo, Bulgaria. Both authors showed that cv. Milena is highly tolerant to all freezing temperatures tested (−18 ºC and −21 °C) after both sufficient and insufficient cold acclimation temperatures. Cluster analysis of cold tolerance test data [Citation51] placed Milena in the clade of the highly tolerant Russian cv. Mironovskaya 808, while Russalka was clustered near to the sensitive Italian spring cultivar San Pastore.
Expression of CBF genes in wheat cultivars with different frost tolerance capacity
High-density mapping strategies, microarray analysis and quantitative RT-PCR have been extensively used to identify possible candidate genes for frost tolerance in both hexaploid and diploid wheat.[Citation16,Citation19,Citation30,Citation54] Among different signalling pathways, the CBF regulon plays a pivotal role in cold acclimation of wheat and other cereal crops. The fine mapping of the Fr-2 locus on chromosome group 5 in wheat revealed a complex structure including multiple CBF genes.[Citation16,Citation19,Citation30,Citation37] According to Badawi et al.,[Citation28] the cluster of CBF genes at Fr-2 loci in hexaploid wheat includes CBF-2, -3, -4, -6, -9, -10, -12, -14, -15, -19, -20, -21 and -22. Knox et al. [Citation41] described that T. monococcum CBF genes at the Fr-2A locus are organized in three subclusters, each composed of 3 or 4 genes: proximal (CBF-2, -4, -9 and -17); central (CBF -14, -15, -12) and distal (CBF -16, -13, -3 and -10). These authors suggested that the locus for frost tolerance is linked to the central gene subcluster (CBF-14, -15, -12) and CBF-6, -19, -20, -21 and -22 do not belong to the frost tolerance gene cluster. Studying the transcription profiles of different T. aestuvum CBF genes using a quantitative PCR system, Vágújfalvi et al. [Citation30] have found that most of the genes at this locus are induced by cold treatment at 2 °C for 2 h, with TaCBF-14, -15 and -16 having the highest transcript levels. These data have been confirmed later by Båga et al. [Citation16], who reported that during cold acclimation TaCBF-14 and -15 are expressed at higher level in the cold tolerant Norstar than in the sensitive winter Manitou.
In this study two winter cultivars (Milena and Russalka) selected for their distinct FT () and the spring cultivar CS were chosen to study the expression pattern of four TaCBF genes during cold acclimation at 2 °C, using qRT-PCR analysis. The four TaCBF genes (CBF15-5AL, CBFA19-5AL CBFA2-5AL and CBFD21.1-5DL), belonging to the groups CBFIIId, CBFIVa and CBFIVb, were chosen on the basis of published information, suggesting their importance in the development of FT in wheat.[Citation28]
The expression results from qRT-PCR () showed that TaCBF genes are induced by LT and, with the exception of CBFA19, attained maximum level of expression between the second and fourth hours of acclimation at 2 °C. Following the kinetics of the transcription, the CBF genes studied here demonstrated different patterns of relative transcript abundance between the highly tolerant and the susceptible winter cultivar as well as between the two winter cultivars and CS.
Figure 2. Quantitative RT-PCR expression analysis of TaCBF genes in highly tolerant winter cultivar Milena, susceptible winter cultivar Russalka and the frost sensitive spring cultivar CS. TaCBF15 (A); TaCBF19 (B); TaCBFA2 (C); TaCBF21 (D). Time points shown are 0 h (control condition at 22 °C); 30 min, 1 h, 2 h, 4 h, 7 h (LT stress at 2 °C); and 24 h (de-acclimation).
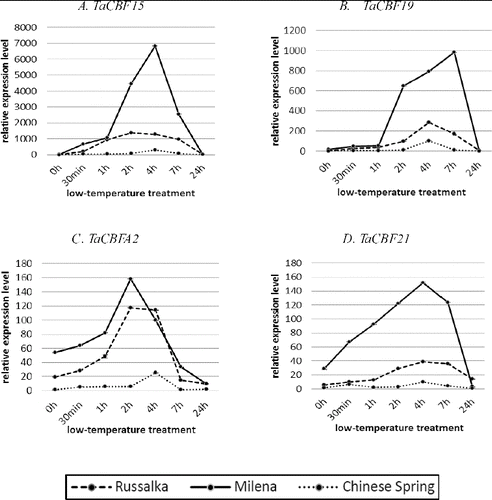
The qRT-PCR analysis revealed that during cold acclimation the expression level of the group IIId genes (CBF15-5AL, CBFA19-5AL) was more pronounced than that of the group IVa (CBFA2-5AL) and IVb ones (CBFD21.1-5DL), although higher transcript accumulation of all studied genes was associated with the higher capacity of the winter cultivars to develop LT tolerance ().
Among the CBF genes analysed in this study, the highest level of accumulation was observed for CBF15-5AL with maximum transcript abundance at 4 h of LT treatment in the tolerant winter cv. Milena and earlier, at 2 h, in the susceptible winter cv. Russalka. It is worth noting that the transcript accumulation in Russalka reached a plateau between 2 and 4 h and the decline thereafter was slower than in the other two analysed cultivars. Induction of CBF15-5AL expression with a peak at 4 h was also observed in CS. However, the pattern was different and the transcript abundance in this cultivar was up to 20- and 50-fold lower than that in Russalka and Milena, respectively ((A)). Similar results for the tolerant winter-hardy cultivar Mir808 have been obtained by Motomura et al.,[Citation32] who reported a transient increase in TaCBFA15 expression with a maximum between the fourth and sixth hours of the LT, resembling the pattern obtained for Milena in the present study. On the other hand, our results for CS disagree with those of Motomura et al.,[Citation32] who found no increase in the transcript abundance of TaCBF15 in the leaf tissue of this cultivar during LT course treatment at 4 °C. The differences in the expression patterns observed in both studies might be a result of the different acclimation temperatures and/or tissues used. The peak expression level in the tolerant Milena was 5-fold higher in comparison to the highest expression level in the susceptible winter Russalka and 22-fold higher compared to CS, while the difference between the susceptible winter Russalka and CS was only 4.5-fold. Vágújfalvi et al. [Citation30] identified that under cold acclimation at 2 °C CBF genes from the central cluster of the Fr2 locus (TaCBF14, -15 and -16) were characterized with the highest transcript levels and those levels were more than fourfold higher in lines carrying the frost tolerant Fr-A2 allele than in those with a frost sensitive one. Up-regulation of the transcript levels of TmCBF16, TmCBF12 and TmCBF15 at mild cold temperatures (15 °C) has been reported by Knox et al. [Citation41] only in the frost tolerant wheat G3116 but not in the frost sensitive Dv92. In contrast, no expression of TaCBF15 has been identified in the crown and leaf tissues of winter tolerant (Harnsek and Solstice) and spring (Paragon) cultivars under both prolonged acclimation and shock experiments in a microarray analysis,[Citation4] which implies that in different wheat genetic backgrounds distinct CBF genes or CBF expression levels are responsible for the development of different levels of freezing tolerance.
High level of expression was also observed for TaCBFA19, although the extent of accumulation of its transcripts was lower than that of TaCBFA15 ((B)). The expression of TaCBFA19 was strongly induced in the tolerant cultivar Milena at the first hour and continued to increase up to seven hours. In contrast, the susceptible winter cultivar Russalka showed a much slower increase in transcript abundance after the first hour with a peak at the 4th hour and a decrease thereafter. Expression in the spring cultivar CS followed the pattern of Russalka. The peak level in the tolerant Milena was 3.5-fold higher in comparison to the highest expression level in the susceptible winter cv. Russalka. Both winter cultivars showed higher maximum expression level, 9.45-fold and 2.7-fold, respectively, as compared to CS. Our results are in accordance to those of Sutton et al.[Citation54] who found that TaCBF19 is differentially expressed between frost tolerant and frost susceptible mutant winter lines of wheat. Both results confirm the involvement of this gene in the development of superior frost tolerance. However, they are in contrast to the study of Knox et al.,[Citation41] who identified this gene as not belonging to the frost tolerance gene cluster.
In comparison to TaCBFA15 and -A19, the expression level of the other two genes, TaCBFA2 and TaCBFD21, was much lower. However, the induction of TaCBFA2 was faster in both winter cultivars and peaked at the second hour of the cold acclimation ((C)), while the pattern of its accumulation in CS was distinct with a peak at the 4 h of the cold acclimation. While in the sensitive winter cv. Russalka the transcript abundance remained at the same level up to 4 h of cold acclimation, in the highly tolerant cultivar Milena the expression of TaCBFA2 dropped quickly after the peak at 2 h. On the basis of microarray analysis, Sutton et al. [Citation54] suggested the involvement of TaCBFA2 in the control of expression of a cluster of Cor/Lea genes on 5A. Therefore, the observed differences in the expression patterns in the two winter cultivars analysed in the present study might be related to distinct regulation of downstream genes, leading to differences in their frost tolerance capacity. However, more detailed studies are needed to further test this hypothesis. The maximum expression level of TaCBFA2 in the frost tolerant Milena was 1.35-fold higher than that in the susceptible Russalka and 27-fold higher than that in the spring cv. CS.
The role of TaCBFA2 in LT tolerance was first proposed by Badawi et al.,[Citation28] who, using spring and winter isogenic wheat lines, demonstrated its down-regulation by the presence of dominant Vrn-1 alleles. Later studies [Citation29,Citation41,Citation54] confirmed the involvement of this gene in the cold acclimation and frost tolerance development in wheat and rye. Significantly higher CBF2 and CBF4 transcript levels have been also observed in the more freezing tolerant winter barley cultivar Nure in comparison to the less tolerant spring cultivar Tremois.[Citation27] Differential expression of TaCBFA2 in both crown and leaf tissues between the tolerant winter and spring genotypes under prolonged and LT shock experiments has been also documented in both microarray analyses performed by the group of Winfield et al.[Citation4]
In our study, the qRT-PCR analysis of TaCBFD21 showed maximum transcript accumulation 4 h after the beginning of CA, which declined thereafter in all analysed cultivars. However, its relative transcript accumulation was much faster in the highly tolerant winter cultivar in comparison to both Russalka and CS ((D)). Its expression at 4 h of LT treatment in Milena was about 4-fold higher than in Russalka and 15-fold higher than in CS.
The genetic capacity of winter cultivars to induce higher levels of expression during LT response was also reflected in the constitutive levels measured under control growth conditions at warmer temperatures (22 °C). Since LT is not involved in the regulation of the expression, the higher levels in winter cultivars versus spring is a sign of the different capacities to express CBF genes and is a consequence of the difference in the capacity of winter cultivars to develop FT. Badawi et al. [Citation28] reported a high and more sustained expression for most CBFIIId, CBFIVa, CBFIVb, CBFIVc and CBFIVd genes. In addition, these groups showed higher basal expression in the winter cultivar versus the spring one. In Arabidopsis, the transgenic constitutive over-expression of the CBF genes results in an increase in freezing tolerance even without prior cold acclimation,[Citation55–57] although this also results in a slow-growing dwarf phenotype, as a consequence of interactions between CBF genes and the gibberellins biosynthetic pathway.[Citation58]
In our study, the CBF genes from groups IIId and IVb showed between 5- and 10-fold higher steady-state expression level at 22 °C in the highly frost tolerant cultivar Milena ((A)–(C)) as compared to cv. Russalka and, respectively, more than 12-fold higher steady-state expression level in comparison to the frost sensitive spring cultivar CS. Noticeable steady-state CBFA2 transcript levels (group IVa) were observed in the studied winter cultivars (19-fold and more) in comparison to CS. However, the difference in the induction levels of CBFA2 in winter cultivars was not as obvious as that between them and CS. Sharp differences in mRNAs steady-state levels between the frost tolerant winter cv. Cheyenne and the frost sensitive spring cultivar CS have been reported.[Citation30] This suggests that the fast and/or the higher cold CBF induction might be associated with the differences in the frost tolerance capacity between cultivars. Several reports [Citation15,Citation27–29,Citation31] have also described that some CBF genes are expressed at low levels in non-stressed conditions in barley, wheat and rye and may lead to continual, albeit low accumulation of the target genes.
Our results suggest that the higher TaCBF- A15, -A19, -A2 and -D21.1 expression is potentially associated with the winter cultivar's superior FT development capacity. They also imply that in the different genetic backgrounds, different genes along the CBF cluster might be implicated in determination of cultivar differences in FT. Additional research aimed at achieving more detailed understanding of the genetic cascades and the complex interaction of genes involved in the development of superior LT tolerance in wheat is highly necessitated.
Conclusions
The comparative quantitative expression analysis (qRT-PCR) showed that the four selected CBF genes are induced and expressed at higher levels in the highly frost tolerant Bulgarian winter cultivar Milena as compared to the more susceptible winter cultivar Russalka. The observed differences in the level of transcription activity of the CBFA15.2, CBFA19, CBFD21 and CBFA2 genes closely correspond to the frost tolerance (LT50) of both winter cultivars. The selected genes could be used as potential functional markers for marker-assisted selection of cold tolerant wheat cultivars carrying Fr-2 alleles.
Functional characterization of other CBF genes and alleles and identification of the CBF genes responsible for better FT capacity of wheat as well as the generation of improved haplotypes combining favourable CBF alleles may provide a useful tool to enhance frost tolerance and hardiness in Bulgarian wheat-breeding programmes.
Additional information
Funding
References
- Laudencia-Chingcuanco D, Ganeshan S, You F, Fowler B, Chibbar R, Anderson O. Genome-wide gene expression analysis supports a developmental model of low temperature tolerance gene regulation in wheat (Triticum aestivum L.). BMC Genomics. 2011;12:299. Available from: http://dx.doi.org/10.1186/1471-2164-12-299
- Båga M, Fowler DB, Chibbar RN. Identification of genomic regions determining the phenological development leading to floral transition in wheat (Triticum aestivum L.). J Exp Bot. 2009;60(12):3575–3585. Available from: http://dx.doi.org/10.1093/jxb/erp199
- Kocsy G, Athmer B, Perovic D, Himmelbach A, Szucs A, Vashegyi I, Schweizer P, Galiba G, Stein N. Regulation of gene expression by chromosome 5A during cold hardening in wheat. Mol Genet Genomics. 2010;283(4):351–363. Available from: http://dx.doi.org/10.1007/s00438-010-0520-0
- Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ. Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J. 2010;8(7):749–771. Available from: http://dx.doi.org/10.1111/j.1467-7652.2010.00536.x
- Galiba G, Vágújfalvi A, Chengxia L, Soltesz A, Dubcovsky J. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 2009;176(1):12–19. Available from: http://dx.doi.org/10.1016/j.plantsci.2008.09.016
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet. 1998;97:968–975. Available from: http://dx.doi.org/10.1007/s001220050978
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 2005;138(4):2364–2373. Available from: http://dx.doi.org/10.1104/pp.105.064287
- Snape JW, Sarma RN, Quarrie SA, Fish L, Galiba G, Sutka J. Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica. 2001;120(3):309–315. Available from: http://dx.doi.org/10.1023/A:1017541505152
- Snape JW, Semikhodskii A, Fish L, Sarma RN, Quarrie SA, Galiba G, Sutka J. Mapping frost tolerance loci in wheat and comparative mapping with other cereals. Acta Agric Hungarica. 1997;45:265–270.
- Galiba G, Kerepsi I, Snape JW, Sutka J. Location of a gene regulating cold-induced carbohydrate production on chromosome 5A of wheat. Theor Appl Genet. 1997;95:265–270. Available from: http://dx.doi.org/10.1007/s001220050558
- Cattivelli L, Baldi P, Crosatti C, Di Fonzo N, Faccioli P, Grossi M, Mastrangelo AM, Pecchioni N, Stanca AM. Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Mol Biol. 2002;48(5–6):649–665. Available from: http://dx.doi.org/10.1023/A:1014824404623
- Toth B, Galiba G, Feher E, Sutka J, Snape JW. Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theor Appl Genet. 2003;107(3):509–514. Available from: http://dx.doi.org/10.1007/s00122-003-1275-3
- Galiba G, Quarrie SA, Sutka J, Morgounov A. RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor Appl Genet. 1995;90(7–8):1174–1179. Available from: http://dx.doi.org/10.1007/BF00222940
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132(4):1849–1860. Available from: http://dx.doi.org/10.1104/pp.103.023523
- Kobayashi F, Takumi S, Kume S, Ishibashi M, Ohno R, Murai K, Nakamura C. Regulation by Vrn-1/Fr-1 chromosomal intervals of CBF-mediated Cor/Lea gene expression and freezing tolerance in common wheat. J Exp Bot. 2005;56(413):887–895. Available from: http://dx.doi.org/10.1093/jxb/eri081
- Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN. Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics. 2007;7(1):53–68. Available from: http://dx.doi.org/10.1007/s10142-006-0030-7
- Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Toth B, Hayes PM, Skinner JS, Pecchioni N. Two loci on chromosome 5H determine low-temperature tolerance in a Nure (winter) x Tremois (spring) barley map. Theor Appl Genet. 2004;108(4):670–680. Available from: http://dx.doi.org/10.1007/s00122-003-1468-9
- Francia E, Barabacshi D, Tondelii A, Laido G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet. 2007;115(8):1083–1091. Available from: http://dx.doi.org/10.1007/s00122-007-0634-x
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Genet Genomics. 2003;269(1):60–67. Available from: http://dx.doi.org/10.1007/s00438-003-0806-6
- Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Ann Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. Available from: http://dx.doi.org/10.1146/annurev.arplant.50.1.571
- Thomashow MF. So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 2001:125(1):89–93. Available from: http://dx.doi.org/10.1104/pp.125.1.89
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14(8):1675–1690. Available from: http://dx.doi.org/10.1105/tpc.003483
- Vogel JT, Zarka DG, van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41(2):195–211. Available from: http://dx.doi.org/10.1111/j.1365-313X.2004.02288.x
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high salt stress. Plant Cell. 1994;6(2):251–264. Available from: http://dx.doi.org/10.1105/tpc.6.2.251
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38(6):982–993. Available from: http://dx.doi.org/10.1111/j.1365-313X.2004.02100.x
- Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17(11):3155–3175. Available from: http://dx.doi.org/10.1105/tpc.105.035568
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. Expression levels of barley Cbf genes at the frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 2007;51(2):308–321. Available from: http://dx.doi.org/10.1111/j.1365-313X.2007.0141.x
- Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F. The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complex of cereal CBFs. Mol Genet Genomics. 2007;277:533–554. Available from: http://dx.doi.org/101007/s00438-006-0206-9
- Campoli C, Matus-Cádiz AM, Pozniak JC, Cattivelli L, Fowler DB. Comparative expression of CBF genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genomics. 2009;282:141–152. Available from: http://dx.doi.org/101007/s00438-009-0451-9
- Vágújfalvi A, Aprile A, Miller A, Dubcovsky J, Delugu G, Galiba G, Cattivelli L. The expression of several Cbf genes at the Fr-A2 locus is linked to frost resistance in wheat. Mol Genet Genomics. 2005;274(5):506–514. Available from: http://dx.doi.org/10.1007/s00438-005-0047-y
- Xue GP. The DNA binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J. 2003;33:373–383. Available from: http://dx.doi.org/10.1046/j.1365-313X.2003.01630.x
- Motomura Y, Kobayashi F, Iehisa JCM, Takumi S. A major quantitative trait locus for cold-responsive gene expression is linked to frost-resistance gene Fr-A2 in common wheat. Breed Sci. 2013;63:58–67. Available from: http://dx.doi.org/10.1270/jsbbs.63.58
- Kobayashi F, Ishibashi M, Takumi S. Transcriptional activation of Cor/Lea genes and increase in abiotic stress tolerance through expression of a wheat DREB2 homolog in transgenic tobacco. Transgenic Res. 2008;17:755–767. Available from: http://dx.doi.org/10.1007/s11248-007-9158-z
- Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotech J. 2011;9:230–249. Available from: http://dx.doi.org/10.1111/j.1467-7652.2010.00547.x
- Takumi S, Shimamura C, Kobayashi F. Increased freezing tolerance through up-regulation of downstream genes via the wheat CBF gene in transgenic tobacco. Plant Physiol Biochem. 2008;46:205–211. Available from: http://dx.doi.org/10.1016/j.plaphy.2007.10.019
- Skinner JS, Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol. 2005;59(4):533–551. Available from: http://dx.doi.org/10.1007/s11103-005-2498-2
- Miller AK, Galiba G, Dubcovsky J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol Genet Genomics. 2006;275(2):193–203. Available from: http://dx.doi.org/10.1007/s00438-005-0076-6
- Mohseni S, Che H, Djillali Z, Dumont E, Nankeu J, Danyluk J. Wheat CBF gene family: identification of polymorphisms in the CBF coding sequence. Genome. 2012;55(12):865–881. Available from: http://dx.doi.org/10.1139/gen-2012-0112
- Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca AM, Pecchioni N. Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet. 2006;112(3):445–454. Available from: http://dx.doi.org/10.1007/s00122-005-0144-7
- Skinner J, Szucs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor Appl Genet. 2006;112(5):832–842. Available from: http://dx.doi.org/10.1007/s00122-005-0185-y
- Knox AK, Li CX, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J. Identification of candidate CBF genes for the frost tolerance locus Fr-A(m)2. Plant Mol Biol. 2008;67(3):257–270. Available from: http://dx.doi.org/10.1007/s11103-008-9316-6
- Todorovska E, Kolev S, Ganeva G, Christov N, Vassilev D. Allele variation in loci for adaptive response and yield traits in Bulgarian wheat. Poster session presented at: 2nd International Symposium on Genomics of Plant Genetic Resources. Proceedings; 2010 Apr 24–27; Bologna, Italy; p. 218.
- Todorovska E, Kolev S, Ganeva G, Vassilev D, Kostov K, Rachovska G. Temporal allele variation in loci for adaptive response and plant stature in Bulgarian wheat. Poster session presented at: 3rd Workshop on TritiGen COST Action FA0604:Triticeae Genomics for advancement of Essential European Crops. Proceedings; 2011 May 3–4; Istanbul, Turkey; p. 43.
- Kolev S, Vassilev D, Todorovska E. Allele variation in loci for adaptive response in Bulgarian wheat cultivars and landraces and its effect on heading date. Plant Genet Resour. 2011;9(2):251–255. Available from: http://dx.doi.org/10.1017/S1479262111000475
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet. 2004;109(8):1677–1686. Available from: http://dx.doi.org/10.1007/s00122-004-1796-4
- Fu D, Szücs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genom. 2005;273(1):54–65. Available from: http://dx.doi.org/10.1007/s00438-004-1095-4
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet. 2007;115(5):721–733. Available from: http://dx.doi.org/10.1007/s00122-007-0603-4
- Nagy Z, Galiba G. Drought and salt tolerance are not necessarily linked: A study on wheat cultivars differing in drought resistance under consecutive water and salinity stresses. J Plant Physiol. 1995;145:168–174. Available from: http://dx.doi.org/10.1023/A:1024335515473
- Christov NK, Imai R, Blume Y. Differential expression of two winter wheat alpha-tubulin genes during cold acclimation. Cell Biol Int. 2008;32(5):574–578. Available from: http://dx.doi.org/10.1016/j.cellbi.2007.11.014
- Livak JK, Schmittgen DT. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25(4):402–408. Available from: http://dx.doi.org/10.1006/meth.2001.1262
- Tsenov N, Chamurkliiski P, Petrova T, Panchev E. [Breeding for cold tolerance in winter wheat (T. aestivum L.) at Dobrudzha Agricultural Institute]. Field Crops Stud. 2012;7(1):53--64. Bulgarian
- Ganeva G, Petrova T, Landjeva S, Todorovska E, Kolev S, Galiba G, Szira F, Bálint AF. Frost tolerance in winter wheat cultivars: different effects of chromosome 5A and association with microsatellite alleles. Biol Plant. 2013;57:184–188. Available from: http://dx.doi.org/10.1007/s10535-012-0267-z
- Tsenov A, Petrova D. [Methods for analysis of stress tolerance of wheat and legume crops]. Plant Sci. 1984;21(6):77–86. Bulgarian
- Sutton F, Chen DG, Ge X, Kenefick D. CBF genes of the Fr-A2 allele are differentially regulated between long-term acclimated crown tissue of freeze-resistant and -susceptible winter wheat mutant lines. BMC Plant Biol. 2009;9:34. Available from: http://dx.doi.org/10.1186/1471-2229-9-34
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54(5):767–781. Available from: doi:10.1023/B:PLAN.0000040902.06881.d4
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 over-expression induces COR genes and enhances freezing tolerance. Science. 1998;280(5360):104–106. Available from: http://dx.doi.org/10.1126/science.280.5360.104
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10(8):1391–1406. Available from: http://dx.doi.org/10.1105/tpc.10.8.1391
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20(8):2117–2129. Available from: http://dx.doi.org/10.1105/tpc108058941