Abstract
Aconitum is a medicinal treasure trove that grows extensively on fertile pastures in Xinjiang Province (China); however, its molecular genetic characteristics are still poorly studied. We studied Aconitum kusnezoffii Reichb., Aconitum soongaricum Stapf., Aconitum carmichaelii Debx. and Aconitum leucostomum Worosch, using random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) techniques, to evaluate their genetic relationship and potential medicinal value. Our results showed that A.kusnezoffii Reichb. and A.soongaricum Stapf. have close genetic relationship and cluster together. Polymorphism rates of 97.25% and 98.92% were achieved by using 15 RAPD and 15 ISSR primers, respectively. Based on Nei's gene diversity (H) and Shannon's index (I), the inter-population diversity (Hs) was higher when compared with the intra-population diversity (Hp). Among the three Aconitum populations, the coefficient of gene differentiation (Gst) was 0.4358 when evaluated by RAPD and 0.5005 by ISSR. The genetic differentiation among the three Aconitum populations was highly significant, suggesting low gene flow (Nm). This was confirmed by the estimates of gene flow (Nm = 0.6473 and Nm = 0.4991, based on ISSR and RAPD data, respectively). Comparing the RAPD and ISSR results, the two DNA markers proved similarly effective in the assessment of the genetic characteristics of the studied Aconitum populations and could be used for reliable fingerprinting and mapping in studies on Aconitum diversity in view of Aconitum suitability for development and protection.
Keywords:
Introduction
Aconite is a valuable traditional Chinese herb of the family Ranunculaceae. The root of this plant is usually used for treating rheumatoid arthritis and easing of pain.[Citation1] In recent years, it has been widely utilized in clinical therapy, e.g. lappaconitine extracted from Aconitum sinomontanum for inhibiting ageing and growth of cancer cells [Citation2] or total alkaloids extracted from Acontium flavum Hand-Mazz for anti-inflammation and local anaesthesia.[Citation3]
Genus Aconitum has 10 species and 5 varieties, and grows on fertile pastures in Xinjiang Province of China. The wild populations of this plant are a real treasure trove as a medicinal resource, but its overgrowth has become a grave threat to ranching in recent years, because aconitine is toxic to animals. To reduce the harm to ranches and at the same time enhance the medicinal utilization of aconite is a highly desired goal. In our previous work,[Citation4] we analysed the chromatographic herbal fingerprint data and identified a variety of alkaloid monomers extracted from aconite plants growing in different regions of Xinjiang Province. By comparing the Chinese Pharmacopoeia records for Aconitum carmichaelii Debx. and Aconitum kusnezoffii Reichb. and different Aconitum species from Xinjiang, we found some similarities and differences in the chemical composition. Therefore, further investigation on the genetic relationship between aconite in Xinjiang and common aconite (A.carmichaelii Debx. and A.kusnezoffii Reichb.) would be important for developing its potential medicinal value.
Molecular marker techniques are powerful and valuable tools used in the analysis of medicinal plants. Among the markers, RAPD (random amplified polymorphic DNA) and ISSR (inter-simple sequence repeat) are generally preferred because of their sensitivity, simplicity and cost-effectiveness. Both RAPD and ISSR markers have been successfully applied to detect genetic similarities or differences in various herbs.[Citation5] As these two types of markers amplify different regions of the genome, when applied together, they allow better analysis of genetic identity and variation.
The aim of the present study was to assess the genetic identity between representatives of genus Aconitum in Xinjiang and A. carmichaelii Debx. and A. kusnezoffii Reichb., by RAPD and ISSR markers. The obtained information about the genetic characteristics will be valuable for screening a variety of Xinjiang representatives of genus Aconitum.
Materials and methods
Sample collection and DNA extraction
Fifteen aconite plants belonging to four species were chosen: Aconitum leucostomum Worosch., Aconitum soongaricum Stapf., A. carmichaelii Debx. and A. kusnezoffii Reichb. The details of the accessions and their geographic origin are listed in . The roots of plants were independently harvested, frozen in liquid nitrogen and stored at −80 °C until DNA extraction. DNA was extracted from 100 mg of root material, using a modified Doyle method.[Citation6] Finally, the extracted DNA samples were quantified with a spectrophotometer (Nanadrop 2000, Thermo Scientific) and diluted to 50 ng/μL in Tris-EDTA buffer; then they were stored at −80 °C for further analyses.
Table 1. Aconitum specimens and their geographic origin.
RAPD analysis
For polymerase chain reaction (PCR), 20 ng of genomic DNA were amplified in a volume of 50 μL containing 10X PCR buffer (10 mmol/L of Tris-HCl, pH 8.3; 50 mmol/L of KCl2), 2.5 mmol/L of MgCl2, 0.2 mmol/L of each deoxyribonucleoside triphosphate (dNTP), 0.4 μmol/L primer and 2 U of Taq DNA polymerase, by means of a thermal cycler (MJ-Mini, BioRad, USA). The cycling programme began with an initial 7 min at 95 °C, followed by 45 cycles at 95 °C for 45 s, 34 °C for 30 s and 72 °C for 90 s, plus a final 10 min at 72 °C and storage at 4 °C. Amplification products were separated by electrophoresis in 1.5% agarose gels. A total of 120 single primers (ShengGong Biotechnology Inc, CHN) were used in the PCR programme, and as a result, 15 primers that amplified polymorphic bands were selected. The sequences of the 15 primers are shown in .
Table 2. Primers sequences.
ISSR analysis
ISSR reactions were performed in a volume of 50 μL containing 25 ng of template DNA, 10X PCR buffer (10 mmol/L of Tris HCl, pH 8.3; 50 mmol/L of KCl2), 2.5 mmol/L of MgCl2, 0.2 mmol/L of each dNTP, 0.4 μmol/L primer and 2 U of Taq DNA polymerase, by means of a thermal cycler (MJ-Mini, BioRad, USA). PCR amplification was performed as follows: initial denaturation at 95 °C for 7 min, followed by 45 cycles at 95 °C for 30 s, 48 °C–60 °C for 45 s and 72 °C for 90 s and a final 10 min extension at 72 °C. Amplification products were separated by electrophoresis in 1.5% agarose gels. A total of 100 primers (ShengGong Biotechnology Inc, CHN) were used in the PCR programme, and as a result, 15 primers that amplified polymorphic bands were selected. The sequences of the 15 primers are shown in .
Data analysis
For the two types of molecular markers, several independent samples from each Aconitum species collected from several locations were tested, and only clear, unambiguous and reproducible bands amplified in both cases were considered. The numbers of polymorphic and monomorphic amplification products were determined for each primer for 15 individuals. To avoid taxonomic weighing, the intensity of bands was not taken into consideration, and only the presence of a band was taken as an indicative. The basic parameters for genetic diversity were calculated with the POPGENE application.[Citation7] The polymorphism of amplification products (P), the number of observed alleles (Na), the mean number of effective alleles (Ne), mean Nei's gene diversity index (H), the Shannon index (I) and the level of gene flow (Nm) [Citation8,Citation9] were determined. Inter-populations diversity (Hs), total gene diversity (Ht) and Nei's coefficient of gene differentiation (Gst) [Citation10] were calculated using the POPGENE 32 software: Gst = (1 − Hs/Ht); Nm, estimate of gene flow from Gst, Nm = 0.5 × (1° Gst)/Gst.. The level of similarity among individuals was established as a percentage of polymorphic bands, and a matrix of genetic similarity was compiled using Dice's coefficient.[Citation11] Dendrograms of the genetic relationship among the 15 individuals of Aconitum were generated by applying the unweighted pair-group arithmetic average (UPGMA) method.[Citation12]
Results and discussion
RAPD and ISSR fingerprinting
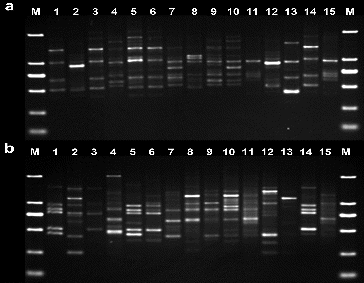
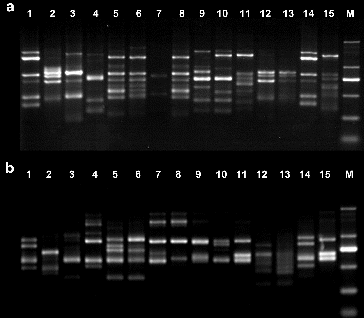
Following screening, 15 primers were chosen for the evaluation of genetic diversity in the four Aconitum species by RAPD and ISSR. The other primers did not generate any amplification products or stable bands. The results from the amplification obtained using the 15 selected primers, the total number of amplification products and the total number of polymorphic fragments, are summarized in . For the 15 Aconitum specimens analysed, a total of 182 bands and a polymorphism index (P) of 97.25% were obtained by RAPD, and a total of 185 bands and a polymorphism index (P) of 99.02%, by ISSR (). The number of bands per primer ranged from 7 to 18, with an average of 12 bands per primer and fragment size ranging from 0.1 to 2 kb in RAPD and ISSR ( and ).
Table 3. Degree of polymorphism for RAPD and ISSR primers in 15 Aconitum specimens.
RAPD and ISSR clustering analysis
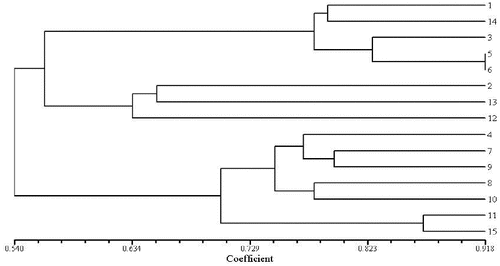
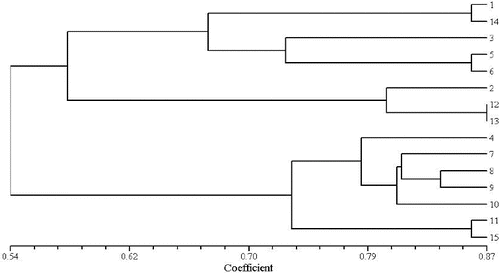
To evaluate the effectiveness of RAPD and ISSR markers for the study of genetic relationships in genus Aconitum, the genetic similarity coefficient was calculated. Cluster analysis was performed for the results obtained by the 15 RAPD and 15 ISSR primers identified to be polymorphic in the 15 studied aconite specimens. UPGMA dendrograms were constructed using SHAN neighbour-joining tree ( and ). The similarity coefficient of the genetic relationships among the 15 Aconitum specimens belonging to three populations was determined to be 0.62 based on the RAPD and ISSR data.
The obtained results revealed that A. kusnezoffii Reichb. and A. soongaricum Stapf. had close genetic relationship and were clustered together. Then, they were clustered with A. carmichaelii Debx. Unlike them, A. leucostomum Worosch. was a unique species, distinct from the other studied Aconitum species with a similarity coefficient of 0.54. RAPD and ISSR clustering analysis ( and ) showed that both techniques have good consistency and repeatability.
Genetic diversity and genetic differentiation
Based on genetic identity, the 15 Aconitum specimens were divided into three clusters in the clustering analysis: P1: A. kusnezoffii Reichb. and A. soongaricum Stapf.; P2: A. carmichaelii Debx.; P3: A. leucostomum Worosch. At the population level, the average values of Na, Ne, H and the percentage of polymorphism were 1.9725% and 1.9892%, 1.5130% and 1.4948%, 0.3072% and 0.3021%, 51.10% and 45.41%, using two molecular markers (RAPD and ISSR), respectively (). The estimates of mean Shannon's index (I) values for the three populations, based on RAPD and ISSR, were similar: 0.4686 and 0.4654, respectively. These high I values at the population level indicate that the populations belong to different species.
Table 4. Mean genetic data of three Aconitum specimens based on RAPD and ISSR markers.
The average coefficient of genetic differentiation (Gst) was 0.4358 and 0.5005, respectively, for RAPD and ISSR, among the three Aconitum populations (). This high genetic differentiation among the three Aconitum populations based on both RAPD and ISSR data, suggested a low level of gene flow. This differentiation was in line with the estimates of gene flow (Nm = 0.6473 and Nm = 0.4991, based on ISSR and RAPD data, respectively).
Table 5. Genetic populations structure and estimate of gene flow within the populations of Aconitum.
These results are interesting, as cultivated Aconitum carmichaeli Debx, and more precisely its lateral roots, is used in one of the traditional Chinese medicine herbs, Radix Aconiti Lateralis Preparata. Due to their wide range of clinical applications, aconite plants have become an important medicine resource. Aconitum kusnezoffii Reichb., A. soongaricum Stapf., A. leucostomum Worosch., Aconitum karakolicum Rapaics and Aconitum leucostomum var. nalatiensis are widespread in several regions in Xinjiang Province.[Citation13] Such medicinal materials are treated as a substitute for the use of A. carmichaelii and A. kusnezoffii, and are used frequently in traditional medicine in the Northwest Territories. However, their quality and purity must meet the market requirements. Aconite products have complex sources, with high-quality medicinal material sometimes mixed together with inferior varieties. It is difficult to classify and genetically analyse them, using traditional morphological characteristics and pedigree analysis. Both RAPD and ISSR technology are valid and useful tools for classification and genetic analysis and have been successfully used for the identification of Aconitum species.[Citation14] These methods can detect a higher level of polymorphism than morphological analysis.
Both RAPD and ISSR are effective techniques for assessment of the genetic diversity of Aconitum spp. medicinal material. They can effectively reveal the genetic relationship between medicinal plant materials. In this way, inter- and intra-species DNA variability can be analysed at the molecular level, which is particularly important in breeding and cultivation of new varieties.
Conclusions
This study is, to the best of our knowledge, the first report on the genetic characteristics of such a range of aconite germplasm in Xinjiang Province. The obtained RAPD and ISSR data indicated high level of genetic differentiation and low gene flow among the studied species. Aconitum leucostomum Worosch. was clustered separately, suggesting that it could be considered as a relatively independent Xinjiang aconite species, because it has farther genetic distance than other species. That is why it may be a promising target for future investigations into its medicinal value. The obtained results are also relevant in view of the suitability of the used sets of RAPD and ISSR primers for development and protection of traditional Chinese medicinal material, and regulation of the Chinese medicinal material market.
Acknowledgements
The authors would like to thank Urumqi OE BioTech. Co., Ltd. for the technical support and data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–186.
- Archak S, Ambika B, Gaikwad D, Gautam EV, Rao VB, Swamy KRM, Karihaloo JL. DNA fingerprinting of India cashew (Anacardium occidentale L.) varieties using RAPD and ISSR techniques. Euphytica. 2003;5:397–404.
- Brijesh S, Daswani PG, Tetali P. Studies on Pongamia pinnate (L.) Pierre leaves: understanding the mechanisms of action in infectious diarrhea. J Zhejiang Univ Sci B. 2006;7:665–674.
- Liu SM, Nie JH, Pan R, Zhao FC. Determination of the content of total alkaloid in Xinjiang genus Aconitum. J Xinjiang Med Univ. 2012;4:1009–5551.
- Bussell JD. The distribution of random amplified polymorphic DNA (RAPD) diversity among populations of Isotoma petraea (Lobeliaceae). Mol Ecol. 1999;8:775–789.
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15.
- Ferrante M, Yeh JTC. Head and flux variability in heterogeneous unsaturated soils under transient flow conditions. Water Resour Res. 2010;5:1471–1479.
- Slatkin M. Gene flow in natural populations. Annu Rev Ecol S. 1985;16:393–430.
- Mcdermott JM, Mcdonald BA. Gene flow in plant pathosystems. Annu Rev Phytopathol. 1993;31:353–373.
- Nei M. Analysis of gene diversity in subdivided populations. PNAS. 1973;70:3321–3323.
- Dice LR. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302.
- Fu YB, Ferdinandez YSN, Phan AT, Coulman B, Richards KW. AFLP variation in four blue grama seed sources. Crop Sci. 2004;44(1):283–288.
- Zhang F, Liu Y, Cai DM. A new variety of Aconitum L. from Xinjiang, China. Northwest Pharm J. 2012;27(6):519–520.
- Luo Q, Ma DW, Wang YH. ISSR identification of genetic diversity in Aconitum carmichaeli. Chinese Traditional and Herbal Drugs. 2006;37(10):1554–1557.