 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
In the present research, statistical Plackett–Burman design (PBD) and central composite design (CCD) were used to optimize the medium's components in order to enhance the chitinase activity of Pseudomonas fluorescens strain HN1205. Among the seven nutritional elements that were studied, crab shell powder, CaCl2·2H2O and yeast extract were proved using PBD to have significant effect on chitinase activity. An optimal medium was obtained by applying a three-factor central composite design, which consisted of crab shell powder (1.0 g/L), CaCl2 (0.5 g/L) and yeast extract (0.5 g/L), with the highest chitinase activity of 1.03 U/mL. This value was 2.87-fold higher than the activity obtained in the lowest productive medium in the design matrix. The chitinase produced by the HN1205 strain was partially purified with 80% ammonium sulphate and anion-exchange chromatography and assessed for inhibition of hatching of nematode eggs. The partially purified chitinase significantly reduced the hatch of Meloidogyne incognita eggs (8.1%), as compared to the effect of 10 mmol/L Tris-HCl buffer (pH 8.0) (49.4%) and boiled chitinase (54.7%). This study demonstrated the role of chitinolytic enzymes produced by P. fluorescens strain HN1205. The obtained optimal activity proved that the enzymes could be potential biological control agents.
Introduction
Chitin, a homopolymer, β-1,4-linked N-acetylglucosamine, is the second most abundant renewable polymer compound in nature, after cellulose. Degradation of chitin is basically catalyzed by chitinases, which are enzymes found in bacteria, fungi, viruses and plants.[Citation1] A large number of microorganisms are a preferred sources of chitinases, because they produce high amounts of these enzymes and it is easy to control their cultural conditions and to follow their harvesting procedures.[Citation2] Chitinases, used in agriculture, are known to carry out functions as biological control agents against root-knot nematode eggs.[Citation3] Meloidogyne incognita is a major plant-parasitic root-knot nematode species, which affects the final yields, production and quality of many annual and perennial crops.[Citation4–6] Although the application of chemical nematicides for controlling the nematodes is usually more effective than other methods, it has caused significant environmental problems that resulted in prohibitions on their use.[Citation7] Because of these harmful effects, the chemical nematicides are losing their popularity among farmers.[Citation6,Citation8] As a result, non-toxic chemical approaches have long been a subject of research in an effort to develop safer alternatives to the use of the nematicides.[Citation9]
Chitinases from microorganisms are a potential weapon for the management of root-knot nematodes, because the nematode eggshell cuticle is composed of a chitin layer,[Citation10] which can be degraded by chitinases. Moreover, the eggshell cuticle is an infection site for microorganisms, which are applied for biological control of nematodes. A number of studies have shown the importance of these enzymes. Chitinase, secreted by Paenibacillus illinoisensis KJA-424, degraded the eggshells of M. incognita, thus, egg hatch was inhibited.[Citation3] In addition, Pochonia chlamydosporia also produces chitinase to penetrate the nematode eggshell during infection.[Citation11] Furthermore, proteolytic enzymes were suggested as nematoxic agents against root-knot nematodes. The cuticle of the nematode's body, which is mainly composed of proteinaceous membranes, can be hydrolyzed by bacterial proteases, peptidyl–peptide hydrolases, gelatinases and collagenases, working as virulence factors against nematodes.[Citation12] This is the reason why research works for new strong chitinolytic organisms and organisms with increased production of enzymes are still of interest.
The genus Pseudomonas is one of the most diverse Gram-negative bacterial genera, isolated from sources ranging from plants to soils and water. The members of this genus are straight or slightly curved rods, motile by means of polar flagella.[Citation13–18] Pseudomonas species have been found to exhibit chitinolytic activity in batch growth cultures.[Citation19–21] Pseudomonas fluorescens BL915 and CHA0 produce extracellular chitinase.[Citation11] P. fluorescens is inhibitory to agricultural pathogens by producing antibiotics and cell wall degrading enzymes, such as β-1,3-glucanase, chitinase and protease.[Citation22] Although chitinase production has been reported in Pseudomonas species, there is no information in previous reports for the statistical optimization of the medium's components for the production of this enzyme.
Optimization of the parameters by statistical experimental design methods can reduce the limitations of the single factor optimization process.[Citation23] The Plackett–Burman design (PBD) is well established and widely used in the statistical experimental designs for screening of medium components as a two-level fractional factorial design.[Citation24] This design is useful in screening studies that uses only k + 1 treatment combinations to estimate the main effects of k factors independently, assuming that all interactions are insignificant.[Citation23,Citation25,Citation26] A saturated design is used in the early stages of scientific tests in order to sort out unimportant factors from a large number of possible factors. When the number of factors in full factorial designs increases, the number of experiments becomes unmanageable.[Citation27] Therefore, PBD comes to be a method for primary screening of medium components.[Citation23] The variables screened by fractional factorial design may be optimized by statistical and mathematical methods, such as response surface methodology (RSM). This tool allows the relationship between independent variables to be evaluated and the response in an effective experimental design to be predicted.[Citation28] Central composite design (CCD) was extensively used, as a statistical technique, for determining the effect of key factors and their further optimization by a small number of experiments.[Citation29] To date, we have not found any research works on the optimization of medium components of P. fluorescens for chitinase production using PBD and CCD RSM.
We have isolated P. fluorescens strain HN1205 with chitinase activity from Suncheon, Jeollanamdo, Korea. The objective of this study was an attempt to optimize the medium components with statistical optimization strategy, in order to improve the chitinase activity by P. fluorescens strain HN1205 using PBD and CCD. In a further study, partially purified enzyme was used for determination of its effect on inhibition of eggs hatching and degradation of eggs of the root-knot nematode M.incognita.
Materials and methods
Culture conditions and preparation of nematodes eggs
Soil rich in crab shell was collected from the seashore at Suncheon, Jeollanamdo, Korea, for the isolation of chitinase-producing bacteria. We made serial dilutions of the collected soil with sterilized water. Inoculations were made on chitin medium containing 250 ml 2% swollen chitin, 2 g/L Na2HPO4, 1 g/L KH2PO4, 5 g/L NaCl, 1 g/L NH4Cl, 0.5 g/L MgSO4·7H2O, 0.5 g/L CaCl2·2H2O, 0.5 g/L yeast extract and 20 g/L agar at pH 7.0. After incubation period of five days at 30 °C, several colonies with wide clear zone on the chitin medium were selected and further screened for their antifungal activities. One of the strains with antifungal activity (data not shown) was identified as P. fluorescens by 16S rRNA sequence analysis and it was designated as P. fluorescens strain HN1205, and maintained in 25% glycerol solution at −70 °C. The GenBank accession number for the nucleotide sequence is HQ610446. The strain was routinely grown in Luria-Bertani medium. The composition of the unoptimized medium, used for chitinase production by P. fluorescens strain HN1205, is shown in . P. fluorescens strain HN1205 was inoculated in a 250 mL Erlenmeyer flask and cultivated at 30 °C with shaking (140 rpm). After four days of incubation, the bacterial culture was used for determination of chitinase production in in vitro experiments.
Table 1. Medium components and experimental variables at different levels, studied in the Plackett–Burman design.
Eggs of the root-knot nematode M.incognita were collected from roots of tomato plants (Solanum lycopersicum L.; Cupirang; NONGWOOBIO CO., LTD), which were previously infected with nematodes in the glasshouse. The plants’ light period was 16 h at 25 °C and they were watered manually once a day. The isolation was performed by placing the roots in 0.5% NaOCl solution and shaking for 3 min.[Citation30] The eggs were collected in a 25 μm sieve. The surface of the eggs was sterilized by exposing it to 0.01% streptomycin sulphate (Sigma) for 1 h before use.
Chitinase activity assay
The bacterial culture broth was incubated for four days and after the incubation period was centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was then used for chitinase activity assay; 0.05 mL of the supernatant was added to 0.45 mL of 50 mmol/L acetate buffer (pH 5.0) and mixed with 0.5 mL of 0.5% colloidal chitin. The reaction mixture was incubated at 37 °C for 60 min and then 0.2 mL of 1 mol/L NaOH were added to terminate the reaction. The mixture was then centrifuged at 10,000 × g for 10 min. After this, 0.75 mL of the supernatant was mixed with 1 mL of Schales' reagent (0.5 mol/L sodium carbonate and 1.5 mmol/L potassium ferricyanide) and boiled for 15 min. The amount of the produced reducing sugars in the supernatant was determined by the following method.[Citation31] Chitinase activity was measured at 420 nm using a spectrophotometer (UV-1650PC, SHIMADZU) and calculated by a standard curve, based on different concentrations of N-acetyl-glucosamine (Sigma). One unit (U) of chitinase activity was defined as the amount of enzyme that liberated 1 μmol/L of N-acetyl-glucosamine per hour.
Statistical screening for medium components
We used the Plackett–Burman screening method with a two-level factorial design for the identification of important medium components with consideration of their main effect. The variables that had the most significant effect, required for maximal chitinase production, were investigated by screening k variables in k + 1 experiments. Each variable was examined at three levels: −1 (low level), 0 (middle level) and +1 (high level).[Citation23] Seven components (variables k = 7) () were selected in this study for each variable. The components were prepared in 250 mL flask containing 100 ml of media (crab shell powder (Purne, Korea), sucrose (Backsul, Korea), KH2PO4, (NH4)2SO4, MgSO4·7H2O, CaCl2·2H2O and yeast extract) according to the design. represents the investigated factors, as well as the levels of each factor, used in the experimental design, whereas shows the design matrix. JMP software trail version (SAS Institute Inc.) was used to analyse the experimental PBD.
Table 2. Plackett–Burman experimental design matrix.
Optimization of the screened ingredients using central composite design (CCD)
The RSM, using a CCD, was applied to optimize and analyse the levels of three important factors and the interaction effects between various medium components, which had a significant influence on chitinase production.[Citation32] The significant factors, identified by the two-level fractional factorial design, were crab shell powder, CaCl2·2H2O and yeast extract. In this study, the experimental plan consisted of 16 trials and the independent variables were coded to 3 different levels: low, middle and high (). All the experiments were done in triplicate and the average chitinase production obtained was taken as the dependent variables or responses. The data obtained from CCD on chitinase production and second-order polynomial equation was then fitted with the JMP software trail version by multiple regression procedure. For a three-factor system, the model was represented by the following quadratic equation:
(1)
(1) where Y is the predicted response in chitinase activity, β0 is the intercept, βi, βii and βij are linear, quadratic and interactive coefficients, respectively, and xi, xj are the coded independent variables. The responses obtained were subjected to a multiple non-linear regression analysis to obtain the coefficients. Estimated coefficients with levels higher than 95% (P < 0.05) were included in the final models.
Table 3. Identification of important variables for chitinase production by Pseudomonas fluorescens strain HN1205 using the Plackett--Burman design.
Partial purification of chitinase produced by P. fluorescens strain HN1205
The P. fluorescens strain HN1205 was grown in the optimized medium at 30 °C for five days on a rotary shaker (140 rpm). After five days the bacterial cells from the culture broth were removed by centrifugation at 6,000 × g for 20 min. The resultant was precipitated by addition of 80% saturated ammonium sulphate and then collected. The precipitate was dialyzed using cellulose tubing (33 mm × 21 mm, SIGMA) with 10 mmol/L Tris-HCl buffer (pH 8.0, buffer T) overnight to desalt and then re-suspended in buffer T. The 10 mL dialysate, containing 11 mg proteins, was loaded onto a diethylaminoethyl (DEAE)-Sepharose fast flow column (2.2 cm × 20 cm, Sigma), pre-equilibrated with buffer T and washed with buffer T. After this, a stepwise elution was made with a gradient of NaCl (0 mol/L to 0.5 mol/L) at a flow rate of 24 mL/h. Two hundred fractions, which were 5.0 mL each, were collected and tested for chitinase activity. The fractions (number 16 to 19) with high chitinase activity were pooled and further concentrated by dialysis as above. Then, we measured the effect of the concentrated sample (2 mL containing 3 mg of proteins; 5.2 U/mL) on egg hatching and degradation of eggshells.
Inhibition of egg hatching and degradation of nematodes eggshells
To estimate the egg hatching rate, 0.01 mL of egg suspension (an average of 50–100 eggs) was taken into 1.5 mL eppendorf tube along with 0.05 mL of partially purified chitinase (5.2 U/mL). The same amount of chitinase, heated for 20 min at 100 °C, or 10 mmol/L Tris-HCl buffer (pH 8.0) solution, were used as controls. The tubes were incubated at 26 °C in the dark for five days. After this period, hatched juveniles from the eggs were counted under a stereomicroscope (Olympus SZX16) with 50× magnification. To estimate the degradation of nematodes eggshells by the chitinase, the same samples were used and photographs were taken under the microscope with camera (FOculuse, F14, Germany). All experiments were performed with three replications.
Results and discussion
Selection of important medium components using Plackett–Burman design
A total of seven variables were identified as important medium components on chitinase production, using the PBD. The designed matrix selected for the screening of significant variables for chitinase production efficiency is shown in . The fittingness of the model was calculated and the variables evidencing statistically significant effects were screened via Student's t-test for analysis of variance (). Factors having a confidence greater than 90% (Prob. > |t| 0.1) were considered to have a significant effect on the response and were further optimized. The lowest p values indicate the most significant factors on chitinase production. X7 (yeast extract) was determined to be the most significant factor (p < 0.005), followed by X1 (crab shell powder) and X6 (CaCl2·2H2O) (p = 0.06 and p = 0.08, respectively). Yeast extract source had a significant effect on chitinase production.[Citation26,Citation33,Citation34] Shrimp shell waste containing chitin source was reported to be an important factor for chitinase production.[Citation35] Gohel et al. [Citation36] has reported CaCl2 as an influential media constituent for chitinase production by Pantoea dispersa. X2 (KH2PO4), X3 ((NH4)2SO4), X4 (MgSO4·7H2O) and X5 (sucrose) are below 90% and therefore considered to be insignificant variables.[Citation37,Citation38] Chitinase production from Streptomyces was inhibited by increased phosphate.[Citation39] The above results showed that the Plackett–Burman experimental design is a powerful tool for screening factors, which have significant effects on chitinase activity.[Citation40]
Table 4. Variables and coded levels of independent variables in central composite design.
Optimization of medium components by CCD
CCD was used to determine the optimal level of the three selected variables (crab shell powder, CaCl2·2H2O and yeast extract) for the chitinase production. A total of 16 trials with different combinations of the three selected variables were performed. shows the coded and actual variable levels with CCD experiments, as well as the chitinase production response. The values of model F and model Prob. > F were found to be 5.10 and 0.0301, respectively, which implies that the model is significant. Value of lack of fit F and lack of fit Prob. > F were found to be 0.05 and 0.9931, respectively, which implies that the lack of fit is insignificant (). The value of correlation coefficient (R2) is 0.8846 indicating a good agreement between actual and predicted values of chitinase production. The value of adjusted R2 (0.7114) for EquationEquation (1)(1)
(1) indicates that 71% of the total variation for chitinase activity is due to the independent variables and only about 29% of the total variation cannot be explained by the model. The value of lack of fit was insignificant (p = 0.9931). The RSM analysis for the optimization of medium constituents showed that chitinase production (Y) by P.fluorescens HN1205 was a function of the concentrations of crab shell powder (X1), CaCl2 (X2) and yeast extract (X3). The following second-order polynomial equation was found to adequately represent the chitinase production:
(2)
(2)
Table 5. Central composite design matrix with experimental and predicted values of chitinase production.
Table 6. Summary of fit, analysis of variance and lack of fit for predictive equation for chitinase production by Pseudomonas fluorescens strain HN1205.
The statistical significance of EquationEquation (2)(2)
(2) was checked by an F-test and by the analysis of variance for the response surface model (). Three-dimensional response plots (JMP software) are shown in to help visualize the effects of crab shell powder, CaCl2 and yeast extract on chitinase production. The plots represent the interaction of two variables, while keeping the third one constant. The response surface shows a curvature along the crab shell powder and CaCl2 (), crab shell powder and yeast extract () and CaCl2 and yeast extract axes (). This may be due to the statistical significance of the quadratic coefficients of these variables in the model. The convex and concave shapes of the plot show that we can find an optimum value for the response in the range of the studied variables. According to the program, a maximal activity of chitinase (1.03 U/mL) can be obtained from a medium containing; crab shell powder (1.0 g/L), CaCl2 (0.5 g/L) and yeast extract (0.5 g/L). This optimum medium has a 2.87-fold higher yield of chitinase activity than that of the less productive medium in the design matrix.
Figure 1. Response surface plot of chitinase production of Pseudomonas fluorescens strain HN1205 as a function of crab shell powder and CaCl2 (a), crab shell powder yeast extract (b) and CaCl2 and yeast extract (c).
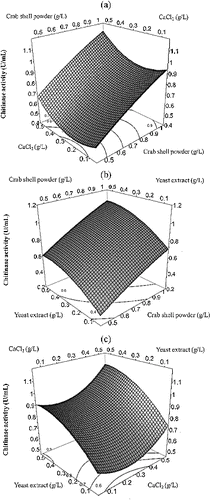
Similar results showed that the addition of yeast extract in the medium stimulated chitinase production by Serratia marcescens [Citation41] and Trichoderma harzianum.[Citation42] Increase in chitinase production by Paenibacillus sabina strain JD2 was shown in medium with CaCl2 presence.[Citation38] The chitinase production was enhanced 4.21-fold by using a statistical medium optimization method in P. dispersa and 4.33-fold where purified chitin compound was used.[Citation36,Citation43] In this study, the 2.87-fold increase in chitinase activity indicated that the PBD and CCD RSM were useful tools for screening factors and optimizing medium components for chitinase production by P. fluorescens strain HN1205. The direct utilization of chitin-containing marine crustacean waste and the bioconversion of shell fish chitin waste has been recently investigated for the production of chitinases.[Citation44] In this study, we used crab shell powder without any chemical processes for chitin purification. Therefore, the use of crude powder resulted in lower chitinase activity, compared to previous studies, which used purified chitin. However, the crude powder reduced the production cost and made it simple to produce microbial chitinases.
Inhibition of egg hatching and degradation of nematodes eggshells
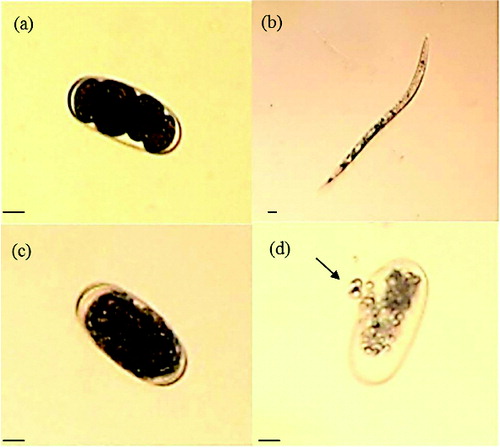
The inhibition of egg hatching by treatment with partially purified chitinase was observed after five days of incubation. Hatching in the presence of chitinase (8.1%) was significantly decreased, as compared to that in 10 mmol/L Tris-HCl buffer (pH 8.0) (49.4%) and boiled chitinase (54.7%) (). The destruction of eggshell structures by the chitinase was clearly demonstrated under the light microscope ().[Citation45] As a result of this experiment, the chitinase inhibited egg hatching and degraded nematodes eggshells, which are composed of chitin. This may be the reason why the chitinase reduced the egg hatching. The exposure of nematode eggs to chitinase suppressed the egg hatching in other studies, too.[Citation46] The purified chitinase LPCHI1 degraded the chitinous layer of M. incognita eggs and significantly influenced their development.[Citation47,Citation45] Meloidogyne javanica eggs were swollen when treated with chitinase and the structure of the eggshells was destroyed.[Citation45,Citation48] Purified chitinases produced from the nematophagous fungi Verticillium chlamydosporium and Verticillium suchlasporium showed cuticle damage on the eggs of Globodera pallida, which is a plant pathogenic nematode.[Citation49] The chitinase activity of Lysobacter capsici YS1215 was induced when M. incognita eggs were added as a chitin supplement during the incubation. Therefore, this result shows that the cuticle of the eggs could be degraded by chitinase secreted by microorganisms.[Citation50] Studies concerning the inhibition of egg hatching and degradation of nematodes eggshells show that the application of chitinase in the biological control of plant pathogenic diseases is possible.
Figure 2. Effect of partially purified chitinase on egg hatching of Meloidogyne incognita after two and five days of incubation at 26 °C. Note: Partially purified chitinase (a), partially purified boiled chitinase (b) and 10 mmol/L Tris-HCl buffer solution (pH 8.0) (c).
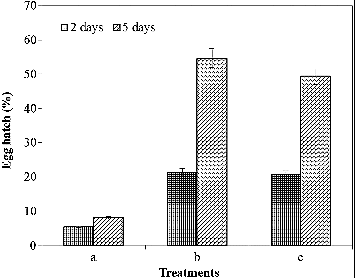
Figure 3. Morphological changes of Meloidogyne incognita eggs caused by partially purified chitinase from P. fluorescens strain HN1205. Note: Eggs exposed only to the buffer at 26 °C for one day (a) and five days (b); on the fifth day in the buffer control, the second stage juvenile (J2) appeared. Eggs treated with partially purified chitinase (5.2 U/mL) at 26 °C for one day (c) and five days (d); the eggs were observed under a light microscope with 200× magnification. An arrow indicates the destruction of eggshell. The scale bars are 20 μm wide.
Conclusions
This study provided results for the elaboration of a suitable medium for the improved chitinase production by P. fluorescens strain HN1205. Maximal chitinase activity was obtained from medium, which contained crab shell powder (1.0 g/L), CaCl2 (0.5 g/L) and yeast extract (0.5 g/L). This result suggested that optimization of the parameters of a medium may be applicable for large scale fermentation processes. Moreover, the study reflects the potential of chitinases for agricultural application (biocontrol of the root-knot nematode M.incognita). Further studies are required to observe the effect of pH and temperature on the enzyme activity and egg hatching inhibition of root-knot nematodes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kao PM, Chen CI, Huang SC, Lin KM, Chang YC, Liu YC. Preparation of fermentation-processed chitin and its application in chitinase affinity adsorption. Process Biochem. 2009;44:343–348.
- Mishra P, Kshirsagar PR, Nilegaonkar SS, Singh SK. Statistical optimization of medium components for production of extracellular chitinase by Basidiobolus ranarum: a novel biocontrol agent against plant pathogenic fungi. J Basic Microbiol. 2012;52:1–10.
- Jung WJ, Jung SJ, An KN, Jin YL, Park RD, Kim YK, Shon BK, Kim TH. Effect of chitinase-producing Paenibacillus illinoisensis KJA-424 on egg hatching of root-knot nematode (Meloidogyne incognita). J Microbiol Biotechnol. 2002;12:865–871.
- Wiratno, Taniwiryono D, Van den Berg H, Riksen JAG, Rietjens IMCM, Djiwanti SR, Kammenga JE, Murk AJ. Nematicidal activity of plant extracts against the root-knot nematode, Meloidogyne incognita. Open Nat Prod J. 2009;2:77–85.
- Nguyen DMC, Seo DJ, Nguyen VN, Kim KY, Park RD, Jung WJ. Nematicidal activity of gallic acid purified from Terminalia nigrovenulosa bark against the root-knot nematode Meloidogyne incognita. Nematology. 2013;15(5):507–518.
- Nguyen DMC, Nguyen VN, Seo DJ, Park RD, Jung WJ. Nematicidal activity of compounds extracted from medicinal plants against the pine wood nematode Bursaphelenchus xylophilus. Nematology. 2009;11(6):835–845.
- Harish S, Saravanakumar D, Radjacommare R, Ebenezar EG, Seetharaman K. Use of plant extracts and biocontrol agents for the management of brown spot disease in rice. Bio Control. 2008;53:555–567.
- Pandey R, Karla A, Tandon S, Mehrotra N, Singh HN, Kumar S. Essential oils as potent sources of nematicidal compounds. J Phytopathology. 2000;148:501–502.
- Ntalli NG, Menkissoglu-Spiroudi U, Giannakou I. Nematicidal activity of powder and extracts of Melia azedarach fruits against Meloidogyne incognita. Ann Appl Biol. 2010;156:309–317.
- Wharton D. Nematode eggshells. Parasitology. 1980;81:447–463.
- Lopez-Llorca LV, Duncan GH. A study of fungal endoparasitism of the cereal cyst nematode Heterodera avenae by scanning electron microscopy. Can J Microbiol. 1988;34:613–619.
- Ray S, Reddigarim SR, Jansma PL, Allen R, Hussey RS. Immunocytochemical analysis of the stage-specific distribution of collagen in the cuticle of Meloidogyne incognita. Fund Appl Nematol. 1996;19:71–78.
- Gaffney TD, Lam ST, Ligon J, Gates K, Frazelle A, Maio JD, Hill S, Goodwin S, Torkewitz N, Allshouse AM, Kempf, HJ, Becker JO. Global regulation of expression of antifungal factors by a Pseudomonas fuorescens biological control strain. Mol Plant Microbe Interactions. 1994;7:455–463.
- Showkat S, Murtaza I, Bhat MA, Abid S, Ali A. Hplc based estimation and molecular characterization of 2, 4 DAPG from Pseudomonas fluorescens isolates of Kashmir. Int J Agri Crop Sci. 2014;7(7):411–416.
- Poorni KE, Manikandan A, Geethanjali S, Percy PK. Production of Pseudomonas fluorescens from agricultural wastes and its application in the preservation of selected vegetables. Adv Appl Sci Res. 2011;2(2):156–160.
- Saranraj P, Sivasakthivelan P, Sakthi, SS. Prevalence and production of plant growth promoting substance by Pseudomonas fluorescens isolates from paddy rhizosphere soil of Cuddalore District, Tamil Nadu, India. Afr J Basic Appl Sci. 2013;5(2):95–101.
- Kanimozhi S, Perinbam K. Siderophore production by Pseudomonas fluorescens Lp1 and its application as biocontrol agent. J Pharm Pract Res. 2011;4(9):3175–3179.
- Rekha V. Isolation, purification, characterization and production of pyoverdine pigment from Pseudomonas fluorescens [dissertation]. Tiruchirappalli (TN): Bharathidasan University; 2011.
- Roberts WK, Selitrennikoff CP. Plant and bacterial chitinases differ in antifungal activity. J Gen Microbiol. 1988;134:169–176.
- Nielsen MN, Sorensen J. Chitinolytic activity of Pseudomonas fluorescens isolates from barley and sugar beet rhizosphere. FEMS Microbiol Ecol. 1999;30(3):217–227.
- Brammacharry U, Paily K. Chitinase like activity of metabolites of Pseudomonas fluorescens Migula on immature stages of the mosquito, Culex quinquefasciatus (Diptera: Culicidae). Afr J Microbiol Res. 2012;6(11):2718–2716.
- Ramyasmruthi S, Pallavi O, Pallavi S, Tilak K, Srividya S. Chitinolytic and secondary metabolite producing Pseudomonas fluorescens isolated from Solanaceae rhizosphere effective against broad spectrum fungal phytopathogens. Asian J Plant Sci Res. 2012;2(1):16–24.
- Priya PP, Mandge S, Archana G. Statistical optimization of production and tannery applications of a keratinolytic serine protease from Bacillus subtilis P13. Process Biochem. 2011;46(5):1110–1117.
- Box GEP. Multi-factor designs of first order. Biometrika. 1952;39:49–57.
- Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325.
- Singh AK, Mehta G, Chhatpar HS. Optimization of medium constituents for improved chitinase production by Paenibacillus sp. D1 using statistical approach. Lett Appl Microbiol. 2009;49:708–714.
- Hunter JS. Statistical design applied to product design. J Qual Technol. 1985;17:210–221.
- James ML. Which response surface design is best. Technometrices 1976;18:411–417.
- Dean AM, Voss D. Design and analysis of experiments. New York (NY): Springer-Verlag New York Inc; 2000.
- Hussey RS, Barker KR. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis Rep. 1973;57:1025–1028.
- Lingappa Y, Lockwood JL. Chitin media for selected isolation and culture of actinomycetes. Phytopathology. 1962;52:317–323.
- Box GEP, Wilson KB. On the experimental attainment of optimum conditions. J Royal Statistical Soc Ser B. 1951;13:1–45.
- Feng T, Zhuang H, Ran Y. The application of cyclodextrin glycosyltransferase in biological science. J Bioequiv Availab. 2011;3(9):202–206.
- Marques HMC. A review on cyclodextrin encapsulation of essential oils and volatiles. Flavour Fragrance J. 2010;25:313–326.
- Ghorbel-Bellaaj O, Manni L, Jellouli K, Hmidet N, Nasri M. Optimization of protease and chitinase production by Bacillus cereus SV1 on shrimp shell waste using statistical experimental design. Biochemical and molecular characterization of the chitinase. Ann Microbiol. 2012;62:1255–1268.
- Gohel V, Chaudhary T, Vyas P, Chhatpar HS. Statistical screenings of medium components for the production of chitinase by the marine isolate Pantoea dispersa. J Biochem Eng. 2006;28:50–56.
- Hao Z, Cai Y, Liao X, Zhang X, Fang Z, Zhang D. Optimization of nutrition factors on chitinase production from a newly isolated Chitiolyticbacter meiyuanensis SYBC-H1. Braz J Microbiol. 2012;43(1):177–186.
- Patel B, Gohel V, Raol B. Statistical optimization of medium components for chitinase production by Paenibacillus sabina strain JD2. Ann Microbiol. 2007;57:589–597.
- Smucker RA, Kim CK. Effects of phosphate on Streptomyces griseus chitinase production. In: Zikaki's JP, editor. Chitin, chitosan and related enzymes. New York (NY): Academic Press; 1984. p. 397–406.
- Han Y, Li Z, Miao X, Zhang F. Statistical optimization of medium components to improve the chitinase activity of Streptomyces sp. Da11 associated with the South China Sea sponge Craniella australiensis. Process Biochem. 2008;43:1088–1093.
- Monreal J, Reese ET. The chitinase of Serratia marcescens. Can J Microbiol. 1969;15:689–696.
- Namoothiri KM, Baiju TV, Sandhya C, Sabu A, Szakacs G, Pandey A. Process optimization for antifungal chitinase production by Trichoderma harziamnum. Process Biochem. 2004;39:1583–1590.
- Gohel V, Trivedi S, Vyas PR, Chhatpar HS. Formulation of medium constituents by multiresponse analysis of central composite design to enhance chitinase production in Pantoea dispersa. Ind J Exp Biol. 2004;42:1123–1131.
- Wang SL, Chang WT. Purification and characterization of two bifunctional chitinases/lysozymes extracellularly produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Appl Environ Microbiol. 1997;63:380–386.
- Lee YS, Anees M, Park YS, Kim SB, Jung WJ, Kim KY. Purification and properties of a Meloidogyne-antagonistic chitinase from Lysobacter capsici YS1215. Nematology. 2014;16:63–72.
- Chen J, Moore WH, Yuen GY, Kobayashi D, Caswell-Chen EP. Influence of Lysobacter enzymogenes Strain C3 on Nematodes. J Nemato. 2006;38(2):233–239.
- Gan Z, Yang J, Tao N, Liang L, Mi Q, Li J, Zhang KQ. Cloning of the gene Lecanicillium psalliotae chitinase Lpchi1 and identification of its potential role in the biocontrol of root-knot nematode Meloidogyne incognita. Appl Microbiol Biotechnol. 2007;76:1309–1317.
- Khan A, Williams KL, Nevalainen HKM. Effects of Paecilomyces lilacinus protease and chitinase on the eggshell structures and hatching of Meloidogyne javanica juveniles. Biol Control. 2004;31:346–352.
- Tikhonov VE, Lopez-Llorca LV, Salinas J, Jansson HB. Purification and characterization of chitinases from the nematophagous fungi Verticillium chlamydosporium and Verticillium suchlasporium. Fungal Genet Biol. 2002;35:67–78.
- Lee YS, Nguyen XH, Naing KW, Park YS, Kim KY. Role of lytic enzymes secreted by Lysobacter capsici YS1215 in the control of root-knot nematode of tomato plants. Indian J Microbiol. 2015;55(1):74–80.
