Abstract
Eukaryotic translation initiation factor 1 family genes (eIF1, eIF1A and eIF1B) play important roles in the 43S complex formation during the mRNA translation of protein synthesis. In particular, eIF1A is stabilizing the binding of Met-tRNA to the 40S ribosomal subunit, promoting the mRNA binding and preventing the premature association of the 40S ribosomal subunit to the 60S ribosomal subunit. eIF1 helps the mRNA scanning for the location recognition of the AUG initiation codon. However, little is known for another eIF1 family gene – eIF1B, especially in plants. In this work, we collected a high salt-tolerant grass Leymus chinensis (Trin.) as the object of our study. We cloned and sequenced the eIF1 family genes from this species. Based on the DNA sequence alignment and the analysis of existing sequences in NCBI’s GenBank database, effective primer sets, specific for L. chinensis, were newly designed. Using these primer sets, the full-length cDNAs of corresponding eIF1 family genes were obtained, and we had submitted these sequencing results to NCBI's GenBank database. The basic local alignment search tool result showed that our sequences were highly identical to relevant existing eIF1 family genes in NCBI's GenBank database. Combined with existing sequence sources, we also constructed phylogenetic trees to further understand the relationship between eIF1 family genes of L. chinensis and other species. This work would provide more eIF1 sequence sources and will lay the foundations for studying and further understanding of the eIF1 gene functions in stress tolerance mechanisms.
Introduction
Soil salinity, most commonly caused by high NaCl concentrations, is one of the most significant abiotic stresses for cultivated plants.[Citation1,Citation2] Much damage could be caused by elevated NaCl concentrations in plants, such as water deficit, ion toxicity, nutrient imbalance and oxidative stress.[Citation2–7] Many evidences suggest that the protein translation and synthesis, which are important processes in plant growth, are very sensitive to NaCl. The start of the protein synthesis is the translation initiation. It includes the binding of a specific initiator methionyl-tRNAi (Met-tRNAi) to the small ribosomal subunit and the formation of a ribosomal initiation complex. In addition to this step, the initiation codon should be located and a translation-competent ribosome should be generated with the joining of the large ribosomal subunit.[Citation8] During these processes, more than 12 protein factors participate, which are called translation initiation factors (eIFs). They all play very important roles by interacting with the ribosomal subunits.[Citation9–11]
The initiator Met-tRNAiMet, eIF2 and GTP form a ternary complex, which is the first step. After this, the ternary complex, together with the multisubunit eIF3, bind to the 40S ribosomal subunit and form a 43S pre-initiation complex.[Citation12] In this process of 43S complex formation, eIF1A plays a stimulating role [Citation13] by stabilizing the binding of Met-tRNA to the 40S ribosomal subunit, promoting the mRNA binding and preventing the premature association of the 40S ribosomal subunit to the 60S ribosomal subunit.[Citation14] The scanning of the mRNA and the location recognition of the AUG initiation codon require eIF1A and factor eIF1.[Citation15,Citation16] In the absence of eIF1, the 40S subunit does not reach the initiation codon accurately in an in vitro reconstituted translation system.[Citation15] eIF1 is considered to play a principal role in the initiation codon selection.[Citation17] This factor also discriminates against assembly of 48S complexes on AUG triplets located only 1–4 nucleotides from the 5′-end of mRNA.[Citation17,Citation18] The role of the other eIF1 factor -- eIF1B is not very well known.
Recently, the overexpression of eIF1A, isolated from salt-tolerant sugar beet, has been found to decrease salt sensitivity and increase salt tolerance in yeast and Arabidopsis thaliana.[Citation2] In a screen for genes regulated by salt stress, the eIF1 was found to be induced by salt stress in Porteresia coarctata.[Citation19] The overexpression of eIF1 in rice has been found to have a potential to increase the salt-stress tolerance in transgenic plants.[Citation20] The eIF1 overexpression was accompanied by a reduced accumulation of Na+ in transgenic rice plants. Little information is available in eIF1B gene cloning and characteristic studies. Although eIF1B has been cloned in some organisms, such as Homo sapiens, Salmo salar, Bos taurus, Canis lupus and Xenopus laevis, it has not been reported in plants so far.
Leymus chinensis (Trin.), a perennial rhizome grass in the Gramineae family, has halophytic ancestors and could be a potential source of halotolerance genes for glycophytic crop plants.[Citation10] This species is widely distributed throughout Northern China, Mongolia and Siberia,[Citation21,Citation22] having intrinsic potential to thrive under environmentally high alkaline-sodic soil conditions (pH 8.5 to 11.5). Due to its good characteristics, L. chinensis has been commonly used to protect the soil from desertification in northern China.[Citation10]
In this work, we described cDNA clones, encoding the L. chinensis eukaryotic translation initiation factor 1 family genes, which include LceIF1, LceIF1A and LceIF1B, by comparing the conversed regions on both edges of the open reading frame (ORF) of these genes with eIF1, eIF1A and eIF1B genes of other species. We analyzed them by using bioinformatics. This work would provide more sequence sources and would help for the better understanding of the characteristics of eIF1 family genes in plants, under the exposure to salt stress.
Materials and methods
Plant materials and growth conditions
Mature seeds of the salt-tolerant L. chinensis were obtained from the natural grassland in Siping, Jilin, China. These seeds have been optimized for their environment by many years of natural evolution. Mature seeds were submerged in sterile water at 4 °C for three days and those, sinking to the bottom, were selected and grown in pots containing clay/vermiculite (3/1, v/v). The cultures were grown with the density of one plant per pot under greenhouse conditions at 25 °C with a 16/8 h (day/night) photoperiod and a relative humidity between 45% and 70%. Seedlings that germinated in these conditions were watered daily with Hoagland nutrient solution.[Citation23]
Gene cloning of LceIF1s and sequence alignment
For the eIF1, eIF1A and eIF1B gene amplifications, three primer sets, specific for L. chinensis, were newly designed based on the nucleotide alignment results of relevant eIF1 family genes from other species. The primer names and sequences are shown in . Among these newly designed primers, eIF1-F1 and eIF1-R1 contained the translation start and end codon, respectively, which are underlined in . Using the corresponding primer set, the full-length cDNA of the LceIF1, LceIF1A and LceIF1B genes was isolated by polymerase chain reaction (PCR) and the PCR products were sequenced.
Table 1. Primer names, sequences and annealing temperatures (TMs) for PCR amplifications of the eIF1 family genes.
The nucleotide sequences and the deduced amino acid sequences of the LceIF1 gene were all aligned using the DNAMAN software version 5.0.
DNA extraction and PCR amplification
Genomic DNA was extracted from fresh leaf tissues of wild-type and transgenic plants using the sodium dodecyl sulphate method.[Citation24] PCR analysis of LceIF1, LceIF1A and LceIF1B was carried out with the following method – 0.5 μmol L−1 of each primer, 10 ng DNA template, 200 μmol L−1 of dATP, dCTP, dGTP and dTTP mix, 1.0 U Taq DNA polymerase (Promega, USA) and the corresponding buffer in a total volume of 20 μL. The PCR amplification was performed with a 95 °C cycle for 5 min, followed by 35 cycles of incubation at 95 °C for 30 s, incubation at the corresponding annealing temperatures (TMs) () for 30 s and 72 °C for 60 s, with a final extension at 72 °C for 7 min. The PCR reactions were performed using the ASTEC PC808 PCR detection system (ASTEC, PC808, Japan). The products were analyzed by 1% agarose gel electrophoresis. Photos were taken and analyzed by the MultiDoc-It Digital imaging system (UVP, Cambridge, UK). All PCR products were purified before the DNA sequencing analysis using a QIAquick PCR Purification Kit (QIAGEN, Korea, Cat. No. 28104). The purified PCR products were then sequenced at MACROGEN Advancing through Genomics, Korea (http://dna.macrogen.com/kor/).
RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from fresh leaf tissues of 200 mmol L−1 NaCl-treated plants using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The NanoPhotometer (IMPLEN, UK) was used to determine the RNA concentration and quality.
Each PCR reaction contained 1.0 μL of each 10 μmol L−1 primer () and 2 μg RNA template in a total volume of 20 μL, using the Maxime RT PreMix Kit (Oligo dT Primer, iNtRON Biotechnology, Korea), according to the manufacturer's instructions. The PCR amplification was performed with an initial denaturation at 95 °C for 5 min, followed by 30 cycles of incubation at 95 °C for 10 s, incubation at the corresponding TMs () for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 7 min. RT-PCR were performed using the ASTEC PC808 PCR detection system (ASTEC, PC808, Japan). The RT-PCR products were analyzed by 1% agarose gel electrophoresis. Photos were taken and analyzed by MultiDoc-It Digital imaging system (UVP, Cambridge, UK).
Sequence analysis
To edit and assemble the complementary strands, the software program DNAMAN version 6.0 (Lynnon Biosoft Corporation, USA) (www.lynon.com) was used. Homologous to our sequences were detected and nucleotide sequence comparisons were done with basic local alignment search tool (BLAST) network services against databases (http://www.ncbi.nlm.nih.gov/). The multiple sequence alignment of eIF1 family genes among different plant species were also performed using the DNAMAN version 6.0 software.
Results and discussion
Primer design for eIF1 family gene amplification and gene cloning
To amplify the eIF1 family genes from L. chinensis, specific primer sets were newly designed. The information of eIF1 family genes (including eIF1, eIF1A and eIF1B) was searched in the NCBI database.
For the eIF1 gene, the systematic BLAST searches identified four genes – from A. thaliana (arabidopsis) (accession number: BT000649) and Zea mays (corn) (accession number: AF034944), as dicotyledonous plant models and from Triticum aestivum (bread wheat) (accession number: AF508969) and Oryza sativa (rice) (accession number: AF094774), as monocotyledonous plant models. To compare the similarities between them and to find the conserved sequence regions for primer design, we constructed a sequence alignment, based on the nucleotide sequences (). According to the conserved sequences, primers encompassing the translation start codon and the translation end codon were designed for the LceIF1 gene amplification. Using the newly designed primer sets for the eIF1 gene, the full-length cDNA of the LceIF1 gene was successfully amplified by RT-PCR, and the product was 363 bp in length. The obtained cDNA sequence of LceIF1 was deposited in the NCBI database with the accession number GU565696. The corresponding protein ID is ADD62702. For the eIF1A gene, the systematic BLAST searches identified six genes from Hordem vulgare (barley) (accession number: AK373449), O. sativa (accession number: AJ563657-58), T. aestivum (accession number: AK332835, WHTEIF1A) and Z. mays (accession number: NM001155525) (). For the eIF1B gene, the systematic BLAST searches identified four eIF1B genes – from S. salar (Atlantic salmon) (accession number: BT057513), Danio rerio (zebrafish) (accession number: BC067620), H. sapiens (human) (accession number: BC006996) and Mus musculus (house mouse) (accession number: NM026892) (). We used the above-described method to design specific primers for L. chinensis. Using the newly designed primer sets, the full-length cDNAs of the LceIF1A and LceIF1B genes were successfully amplified by RT-PCR, and the products were 435 and 180 bp in length, respectively. The obtained cDNA sequence of LceIF1A was deposited in the NCBI database with the accession number HM071922. The corresponding protein ID is ADI47121. The obtained cDNA sequence of LceIF1B was deposited in the NCBI database with the accession number HM210727. The corresponding protein ID is ADN77997.
Figure 1. DNA sequence alignment of eIF1 from different plant species.
Note: AteIF1 (accession number: BT000649) from A. thaliana, OseIF1 (accession number: AF094774) from O. sativa, TaeIF1 (accession number: AF508969) from T. aestivum and ZmeIF1 (accession number: AF034944) from Z. mays. The two red parts are the newly designed forward and reverse primers’ binding sites, respectively.

Figure 2. DNA sequence alignment of eIF1A from different plant species.
Note: HveIF1A (accession number: AK373449) from H. vulgare, OseIF1A-1 (accession number: AJ563657) and OseIF1A-2 (accession number: AJ563658) from O. sativa, TaeIF1A-1 (accession number: WHTEIF1A) and TaeIF1A-2 (accession number: AK332835) from T. aestivum, and ZmeIF1A (accession number: NM001155525) from Z. mays. The two red parts are the newly designed forward and reverse primer binding sites, respectively.
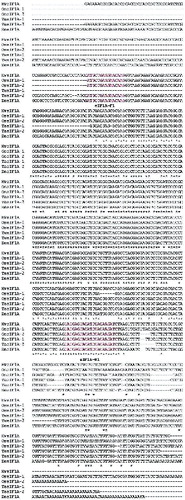
Figure 3. DNA sequence alignment of eIF1B from different species.
Note: SseIF1B (accession number: BT057513) from S. salar, DreIF1B (accession number: BC067620) from D. rerio, HseIF1B (accession number: BC006996) from H. sapiens and MmeIF1B (accession number: NM026892) from M. musculus. The two red parts are the newly designed forward and reverse primer binding sites, respectively.

Sequence analysis of LceIF1 cDNA
The full-length cDNA of the LceIF1 gene isolated with RT-PCR by using the newly designed primers contained an ORF of 348 bp and encoded a protein containing 115 amino acids ((A)). Sequence analysis showed that the cDNA sequence belonged to a member of the eIFs family and the SUI1/eIF1 superfamily. There were some putative RNA binding sites, marked in red, boldface letters, of which DQ were considered as the start sites affecting mutations. In addition, the predicted RNA interaction sites were found and were marked as YSK..K..KKE by blue boldface letters ((A) and 4(B)).
Figure 4. Bioinformatics analysis of L. chinensis eIF1 gene and protein.
Note: (A) Nucleotide and deduced amino acid sequence of LceIF1. The amino acid is indicated by the one letter designation below the nucleotide sequence. The putative binding sites are marked as red, boldface letters. The predicted RNA interaction sites are marked as blue, boldface letters. The mutations affecting start sites are underlined. (B) Multiple sequence alignment among EIF1 protein sequences, including AtEIF1 (accession number: AAN18215) from A. thaliana, OsEIF1 (accession number: AAC67556) from O. sativa, TaEIF1 (accession number: AAM34279) from T. aestivum, ZmEIF1 (accession number: AAB88615) from Z. mays, and LcEIF1 (accession number: ADD62702). (C) Phylogenetic tree analysis among these EIF1 protein sequences.

To further understand the phylogenetic relationship between LceIF1 and eIF1 genes from other plant species, the deduced amino acid sequence of the LceIF1 gene was aligned using the DNAMAN software version 5.0 with existing eIF1 gene protein sequences, including AtEIF1 (accession number: AAN18215) from A. thaliana, OsEIF1 (accession number: AAC67556) from O. sativa, TaEIF1 (accession number: AAM34279) from T. aestivum, ZmEIF1 (accession number: AAB88615) from Z. mays and the deduced amino acid sequence from LcEIF1 (accession number: ADD62702) ((B)). A phylogenetic tree was constructed by using the neighbour-joining method, based on amino acid sequence comparisons. The result revealed that the LcEIF1 protein sequence showed 89% identity with A. thaliana EIF1 (AtEIF1) and 87% identity with T. aestivum EIF1 (TaEIF1), O. sativa EIF1 (OsEIF1) and Z. mays EIF1 (ZmEIF1) ((C)).
Cloning and sequence analysis of LceIF1A cDNA
The full-length cDNA of the LceIF1A gene isolated with RT-PCR by using the newly designed primers contained a full-length ORF of 435 bp, encoding a protein, containing 144 amino acids ((A)). Due to few sequence resources of EIF1A, we only collected A. thaliana and Z. mays as dicotyledonous plant models, and O. sativa as monocotyledonous plant model. To further understand the phylogenetic relationship between LceIF1A and eIF1A genes from other plant species, the deduced amino acid sequence of the LceIF1A gene was aligned using the DNAMAN software version 5.0 with existing eIF1A gene protein sequences, including AtEIF1A (accession number: BAK04646) from A. thaliana, OsEIF1A-1 (accession number: CAD91550) and OsEIF1A-2 (accession number: CAD91551) from O. sativa, ZmEIF1A-1 (accession number: NP001148997) and ZmEIF1A-2 (accession number: ACG49138) from Z. mays and LcEIF1A (accession number: ADI47121) ((B)). The sequence analysis showed that the cDNA sequence shared a high level of nucleotide sequence identity with the EIF1A described in other organisms (98% identical to the EIF1A from Z. mays, 95% identical to the EIF1A from O. sativa ((C)). The ZmEIF1A protein contained 188 amino acids. Except this protein, all of the other EIF1A proteins had similar length of their sequences ((B)). That is why ZmEIF1A-1 was only 91% identical to our protein sequence ((C)). In addition, its amino acid sequence contained all the conserved amino acid residues characteristic of the EIF1A,[Citation8] such as the oligonucleotide-binding site postulated to bind the RNA during the translation initiation ((A)).
Figure 5. Bioinformatics analysis of L. chinensis eIF1A gene and protein.
Note: (A) Nucleotide and deduced amino acid sequence of LceIF1A. The start codon and ending codon are marked red. The conserved oligonucleotide-binding site is underlined. (B) Multiple sequence alignment among EIF1A protein sequences, including AtEIF1A (accession number: BAK04646) from A. thaliana, OsEIF1A-1 (accession number: CAD91550) and OsEIF1A-2 (accession number: CAD91551) from O. sativa, ZmEIF1A-1 (accession number: NP001148997) and ZmEIF1A-2 (accession number: ACG49138) from Z. mays and LcEIF1A (accession number: ADI47121). (C) Phylogenetic tree analysis among these EIF1A protein sequences.

Cloning and sequence analysis of LceIF1B cDNA
The full-length cDNA of the LceIF1B gene isolated with RT-PCR by using the newly designed primers contained the full-length ORF of 157 bp, encoding a protein containing 58 amino acids ((A)). As authors’ knowledge, eIF1B cloning was scarce reported in plants. The reason for this might be that the expression level of eIF1B is so low, that it cannot be successfully amplified by RT-PCR, unless the plant materials are treated with stress factors, such as salt stress. Little information is available in order to compare the results in low similarity rate between our sequence and others. However, fortunately, we successfully amplified the full-length cDNA of the eIF1B gene from L. chinensis using our designed primer sets. To further understand the phylogenetic relationship between LceIF1B and eIF1B genes from other organisms, the deduced amino acid sequence of the LceIF1B gene was aligned using the DNAMAN software version 5.0 with existing eIF1B gene protein sequences, including SsEIF1B (accession number: ACM09385) from S. salar, DrEIF1B (accession number: AAH67620) from D. rerio, HsEIF1B (accession number: AAH06996) from H. sapiens, MmEIF1B (accession number: NP081168) from M. musculus and LcEIF1B (accession number: ADN77997) ((B)). According to the alignment results of the EIF1B protein sequences, large length variations were found: LcEIF1B was much shorter than EIF1B from the other organisms, among which human EIF1B was the longest, with 133 amino acids, and EIF1B from most animals contained 113 amino acids ((B)) (). The difference might be mainly between plants and animals and this situation was also found in other genes. In addition, a phylogenetic tree was also constructed based on the alignment result ((C)). In the phylogenetic tree, we suggested that the protein sequence only shared 45% identity with the EIF1B described in other organisms, such as D. rerio, S. salar, H. sapiens and M. musculus, which was not only caused by sequence variation, but also by length variation. Our sequence provided an important source of eIF1B from plants that would help to further understand the eIF1B gene function.
Figure 6. Bioinformatics analysis of L. chinensis eIF1B gene and protein.
Note: (A) Nucleotide and deduced amino acid sequence of LceIFB. (B) Multiple sequence alignment among EIF1B protein sequences, including SsEIF1B (accession number: ACM09385) from S. salar, DrEIF1B (accession number: AAH67620) from D. rerio, HsEIF1B (accession number: AAH06996) from H. sapiens, MmEIF1B (accession number: NP081168) from M. musculus and LcEIF1B (accession number: ADN77997). (C) Phylogenetic tree analysis among these EIF1B protein sequences.
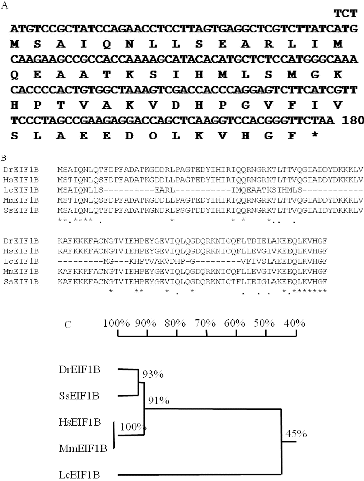
Table 2. Amino acid analysis of protein EIF1B in different organisms.
Conclusions
This work provided not only a simple method for cloning of positive genes, but also additional eIF1 family gene sources. Our results further defined the class and number of eIF1 family genes, especially the eIF1 and eIF1A genes, whose nomenclature and functions are easily confused. Although the eIF1 and eIF1A genes were indeed similar with each other in their functions during the scanning of the mRNA and during the location recognition of the AUG initiation codon, both of the genes had certain differences in their gene type and function. In particular, the eIF1B gene was scarcely cloned from plant materials, so this work filled a gap in the cognition of this gene. With this study, we indicated that the eIF1B gene was one of the stress-regulated genes, because it could be induced by some stresses by showing an overexpression. In a word, this work would provide the basis for further understanding of eIF1 family gene function during the translation initiation or stress tolerance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448.
- Rausell A, Kanhonou R, Yenush L, Serrano R, Ros R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. J Plant. 2003;34:257–267.
- Niu XM, Bressan RA, Hasgawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Phys. 1995;109:735–742.
- Blumwald E. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol. 2000;12:431–434.
- Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71.
- Serrano R. Salt tolerance in plants and microorganisms, toxicity targets and defense responses. Int Rev Cytiol. 1996;165:1–52.
- Serrano R, Gaxiola R. Microbial models and salt stress tolerance in plants. CRC Crit Rev Plant Sci. 1994;13:121–138.
- Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56–63.
- Asano K, Clayton J, Shalev A, Hinebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546.
- Jin CW, Sun YL, Hong SK. Gene cloning of an important eukaryotic translation initiation factor family, eIF2A gene in halophytic Leymus chinensis (Trin.). Biotechnol Biotechnol Equip. 2014;28(4):622–626.
- Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC. Characterization of mammalian eIF2A and identification of the yeast homolog. J Biol Chem. 2002;277:37079–37087.
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556.
- Chaudhuri J, Si K, Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J Biol Chem. 1997;272:7883–7891.
- Battiste JL, Pestova TV, Hellen CUT, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell. 2000;5:109–119.
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–858.
- Pestova TV, Hellen CUT. Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA. 2001;7:1496–1505.
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922.
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17(22):2786–2797.
- Latha R, Salekdeh GH, Bennett J, Swaminathan MS. Molecular analysis of a stress-induced cDNA encoding the translation initiation factor, eIF1, from the salt-tolerant wild relative of rice, Porteresia coarctata. Funct Plant Biol. 2004;31:1035–1042.
- Diédhiou CJ, Popova OV, Dietz KJ, Golldack D. The SUI-homologous translation initiation factor eIF-1 is involved in regulation of ion homeostasis in rice. Plant Biol. 2008;10:298–309.
- Huang Z, Zhu J, Mu X, Lin J. Pollen dispersion, pollen viability and pistil receptivity in Leymus chinensis. Ann Bot. 2004;93:295–301.
- Sun YL, Hong SK. Effects of plant growth regulators and l -glutamic acid on shoot organogenesis in the halophyte Leymus chinensis (Trin.). Plant Cell Tissue Organ Cult. 2010;100(3):317–328.
- Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Berkeley (CA): Taylor & Francis; 1950.
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21.
