Abstract
Rice noodles, a traditional Chinese food, are made using natural fermentation. The fermentation process plays an important role for the texture and flavour, which are due to the metabolism of the microorganisms involved, especially lactic acid bacteria (LAB). However, the fermentation of rice noodles is a non-controlled and spontaneous process; the quality of products is not stable. In order to improve and stabilize the quality of the production, the LAB functions on rice noodles during fermentation need to be elucidated. In this study, the community composition and dynamics of LAB associated with the fermented rice were investigated by using a culture-dependent approach in combination with a culture-independent approach, polymerase chain reaction–denaturing gradient gel electrophoresis (PCR-DGGE). Sixty-four strains were isolated by culture-dependent analysis in different rice noodles factories and mainly 10 species in total were found. In addition, PCR-DGGE detected 17 species (including uncultured species) in total. Seven Lactobacillus species were detected by both methods, namely Lb. delbrueckii, Lb. salivarius, Lb. helveticus, Lb. reuteri, Lb. fermentum, Lb. amylovorus and Lb. oris, and the rest were detected by different methods separately. The potential role of these predominant LAB in the fermentation process is discussed. In general, the two techniques complement each other to give a better understanding on the LAB composition, which is necessary for the isolation of functional strains and improvement of the quality and consistency of rice noodles.
Introduction
Rice noodles, made from white rice and water, are a popular traditional staple food in south China. It originated during the Qin dynasty (259–210 BC) and has been consumed for more than 2000 years in China. Changde (a city in Hunan province) is one of the best-known ‘rice-noodle oriented areas’, with about 300 thousand residents using rice noodles for breakfast and the daily consumption of fresh rice noodles amounting to about 75,000 kg.[Citation1]
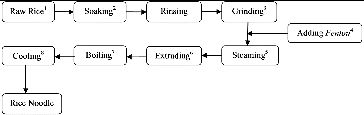
The main stages involved in the production of rice noodles include soaking, grinding (pasting), heating (steaming), extruding, boiling, and cooling ().[Citation2] Based on the duration of the soaking process and whether it is accompanied by fermentation, rice noodles are classified as unfermented noodles and fermented noodles. Soaking is essential for the production of fermented noodles because it increases the water content of the noodles and allows natural fermentation to occur. Fermentation gives rice noodles a better texture and flavour, which is due to the metabolism of the microorganisms involved.[Citation3–6] The process takes 2–3 days in summer and 4–5 days in winter and is dominated by microbes, especially lactic acid bacteria (LAB) and yeasts.[Citation7]
Figure 1. Production process of fresh rice noodles. (1) Old Indica rice is used, normally, milled rice is stored for more than half year after milling. (2) At room temperature and natural environment, treated for 2–4 d. (3) Grinding fermented rice with added water to smooth slurry. (4) Fentou, the beginning and the end processed rice noodle from the day before. About 10% (based on the weight of the rinsed rice) is added. (5) High pressure steam will heat the rice slurry on the belt to partly gelatinized and sticky paste. (6) Molding procedure, round cross section rice noodles are obtained by extruding machine. (7) Extruded noodles go directly into boiling water for 1–2 min. (8) Cooling boiled rice noodle rapidly to 24 °C –26 °C in a trough filled with tap water.
It is known that the cereal fermentation relies on the activity of LAB (e.g., Lactobacillus and Pediococcus spp.), Enterobacter spp., yeasts (e.g., Candida, Debaryomyces, Endomycopsis, Hansenula, Pichia, Saccharomyces and Trichosporon spp.) and filamentous fungi (e.g., Amylomyces, Aspergillus, Mucor and Rhizopus spp.). The fermentation process modifies the taste, flavour, acidity, digestibility and texture of the noodles.[Citation8]
However, the fermentation of rice noodles is a non-controlled and spontaneous process and, that is why, the quality of products is not stable. Therefore, to ensure the quality of rice noodles, deeper knowledge is needed about the microbial population during the fermentation process. Previous studies on rice noodles have mainly used culture-dependent methods; however, these cannot be used to identify some important microbial species, and this might influence the examination of the microbial diversity.[Citation9] Culture-independent methods such as polymerase chain reaction–denaturing gradient gel electrophoresis (PCR-DGGE) are widely applied to study the microbial diversity in a rapid and economical way.[Citation10] These methods accelerate the identification of dominant microorganisms and the development of starter cultures.[Citation9]
In this study, a combination of culture-dependent and culture-independent methods was used to investigate the LAB communities during the fermentation process of rice noodles. Information on the LAB ecology was obtained, which is necessary for the development of well-characterized starter cultures to improve and stabilize the quality of fresh rice noodle products.
Materials and methods
Sampling and storage
Samples of fermented supernatants and fermented rice were collected from three factories. From each factory, the samples were collected at 0, 24, 48 and 72 h during the fermentation process, which lasts for 3 d in summer. All of the samples were collected and placed in sterile glass bottles, then transported to the laboratory in an ice box. For culture-based investigations, the samples were analysed immediately upon arrival, and for culture-independent investigations, the samples were stored at −20 °C prior to further analyses.
Microbial counts and isolation
Culture-based microbiological investigations were conducted at four time points during the fermentation process. A total of 25 g of sample was transferred into sterile conical flasks, to which 225 mL of sodium chloride solution (0.85%) was added. The samples were then homogenized and serially diluted.
Total aerobic bacteria (AB) were scored on plate count agar, with the plates incubated at 30 °C for 1–2 d. LAB were counted using de Man, Rogosa and Sharpe agar medium, with the agar plates incubated at 30 °C for 72 h. Gram-staining and catalase reactions were carried out to confirm the presence of LAB. Gram-positive and catalase-negative organisms were selected as LAB. Fungi (yeasts and molds) were scored using two different media, malt extract agar (MEA) and Rose bengal chloramphenicol agar (RBCA). Then 100 μL of diluted suspension was added and spread onto these plates, which were then incubated at 25 °C for 2–4 d.
Duplicate counts were performed for each of the duplicated samples, and the results are reported as means with standard deviation (±SD; n = 4). Isolates of LAB were randomly selected from plates with 20–300 colonies; the number of isolates is the square root of the total number of colonies.[Citation11]
Identification of the isolates
The isolates were identified and sequenced at the Beijing Center for Physical and Chemical Analysis (Beijing, China). Sequence identity was determined by accessing the nucleotide basic local alignment search tool (BLAST) database of GenBank (http://blast.ncbi.nlm.nih.gov). The 16S rRNA gene sequences of LAB in this study were deposited in GenBank under accession numbers KP317674–KP317737.
DNA extraction and PCR-DGGE analysis
DNA extraction
For culture-independent investigations, total DNA was extracted from each sample by the method described by Wang et al. [Citation12]. The concentrations of DNA samples collected were determined by ultraviolet–visible spectroscopy (Unico, Shanghai, China) and then diluted to 1–50 ng/μL.
PCR-DGGE analysis
The V3 region of the 16S rRNA gene was amplified using primers 338GC-for and 518-rev.[Citation13] The PCR was performed in 50 μL reaction mixtures containing 37.75 μL of ddH2O, 5 μL of PCR buffer, 4 μL of deoxyribonucleoside triphosphates (2 mmol/L), 1 μL of each primer (10 μmol/L), 1 μL of DNA template and 0.25 μL of Ex Taq DNA polymerase (5 U/μL; Takara, Japan). All PCR reactions were carried out in an AG 223B1 Thermoblock (Eppendorf, USA) with the following PCR conditions: initial denaturation of double-stranded DNA for 5 min at 94 °C; 35 cycles each consisting of 30 s at 94 °C, 30 s at 56 °C and 1 min at 72 °C and a final extension of incomplete products for 7 min at 72 °C, followed by cooling at 4 °C. The PCR products were then analysed by 1.5% agarose gel electrophoresis and stored at −20 °C for further sequencing analysis.[Citation11,Citation14]
The amplified products were subjected to the DGGE analysis using the DCode system (Bio-Rad, Hercules, CA, USA) on 20 cm × 16 cm × 1 mm gels. Electrophoresis was performed at 60 °C in 0.5× TAE buffer (20 mmol/L Tris-acetate, 2 mmol/L ethylenediaminetetraacetic acid; pH 8.0) using 8% polyacrylamide gels containing a 30%–60% urea-formamide linear denaturing gradient (100% corresponds to 7 mol/L urea and 40% (v/v) formamide). Samples were run for 16 h at 85 V.[Citation11,Citation14]
Following electrophoresis, the gels were stained with the AgNO3 solution as follows. The gel was fixed and shaken gently in 1 × Cairn's fixation solution (200 mL 96% ethanol; 10 mL acetic acid; 40 mL demi-water) for 3 min. The gel was transferred to a freshly made AgNO3 staining solution (2 g/L of AgNO3), shaken gently for 10 min and then briefly rinsed in water. The stained gel was developed in a freshly made developing solution (10 mg NaBH4, 250 mL 1.5% NaOH solution, 750 μL formaldehyde) until the desired exposure was achieved. Selected bands were excised from the gel, put in sterile water and boiled for 30 min to release DNA. The gel solutions were cooled and stored overnight at 4 °C.[Citation11,Citation14]
Sequencing of DGGE bands
The DNA obtained as supernatants was re-amplified using the above-described primers without a GC clamp. The PCR products were sequenced at the Beijing Center for Physical and Chemical Analysis (Beijing, China). Sequence identity was determined by accessing GenBank by using the nucleotide BLAST program. The sequences of excised bands in this study were deposited in GenBank under accession numbers KP317759–KP317780 for LAB.
Results and discussion
Bacterial and fungal counts during the fermentation process
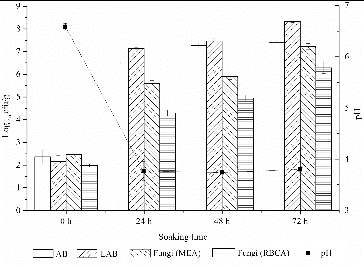
The prevalence of AB, LAB and fungi during the fermentation process is illustrated in , with the pH values at the respective stages indicated. It was observed that all species increased in counts over the whole course of 3 d, which is in agreement with other studies.[Citation7] The AB and LAB counts (scored as colony-forming units) increased dramatically by 7 log cfu/g after 48 h. The LAB counts were higher than the aerobic plate counts during the entire fermentation process. These results indicated that LAB were the dominant microorganisms. The fungal counts increased rapidly during the whole fermentation period, which may be due to the higher temperature and humidity in summer. Isolation using MEA resulted in a higher number of countable colonies in comparison with isolation using other RBCA.
In this study, the pH of the supernatants decreased below 4.0 after 24 h of fermentation and then remained constant at 3.5–4.0. This observation is consistent with a previous report [Citation7] and may be attributed to the increasing number of LAB in the medium. The growth of pathogens, such as coliforms, Staphylococcus aureus and Salmonella spp., is inhibited during fermentation.[Citation15,Citation16] This might imply that fermentation could be an effective method for inactivating contaminating microbes. Thus, the results of the low pH confirmed the safety of the fermented rice noodles.
Analysis of LAB diversity based on 16S rRNA sequences
A total of 64 LAB strains were randomly selected and identified by 16S rRNA gene sequencing. Ten species were identified and most of the sequenced isolates showed high sequence similarity (99%–100%) with GenBank sequences. The results demonstrated that Lactobacillus fermentum predominated during the fermentation process in different factories, representing approximately 38.1%, 45.8% and 68.4%, respectively (). Lb. salivarius and Lb. plantarum were the second predominant microorganisms during the fermentation. The former constituted 28.6% and 15.8%, respectively, in the samples from factory A and C, and the latter accounted for 25.0% in the sample from factory B. Other LAB comprised seven species: Lactococcus garvieae, Enterococcus faecalis, Lb. oris, Lb. reuteri, Lb. delbrueckii, Lb. helveticus and Lb. amylovorus. Similar to previous studies, Lb. plantarum, Lb. fermentum, Lb. delbrueckii and Lb. helveticus were also detected as predominant species.[Citation7] And some LAB which were not detected in this study but are commonly encountered in fermented rice noodles included Bifidobacterium magnum, Lb. rhamnosus and Lb. sakei subsp. sakei. These species play an important role for organic acid production and the flavour of rice noodles during fermentation.[Citation17]
Table 1. LAB composition of fermented rice in different factories by culture-dependent methods.
Currently, the investigation of LAB associations in fermentation foods attracts substantial interest.[Citation14] Recent studies have focused on the preferential growth of particular organisms and the spontaneous succession of microbial communities.[Citation18,Citation19] The changes in LAB throughout the fermentation process are important for the production of rice noodles. A comprehensive understanding of this process will aid in devising strategies to improve the quality and taste of the product. To date, however, the microbiota present during the fermentation process of the rice noodle production has received little attention.
The results demonstrated that LAB play a major role in fermentation, with Lb. fermentum, Lb. salivarius and Lb. plantarum being the dominant LAB present throughout the fermentation process. Several LAB identified in this study are also present in other fermented foods and beverages.[Citation8,Citation20] Some studies have demonstrated that these LAB can convert sugar, malic acid and citric acid into lactic acid and other substances. Because of the acid tolerance, these LAB could still be detected at the end of the fermentation process, and they can change the texture of starch.[Citation21,Citation22] They all showed trypsin and lipase activities,[Citation23] ability to digest proteins, lipids and starch. Meanwhile, the lactic acid produced by LAB could modify the characteristics and improve the sensory properties of rice noodles.[Citation24] The potential to modify and purify the structure of rice starch by means of LAB is beneficial to rice noodle making. Therefore, further investigations about the enzyme activities and lactic acid production of the dominant LAB species could be performed in order to develop a rice noodles fermentation starter with better quality and stability.
DGGE analysis of LAB populations
DGGE analysis of the amplified 16S rRNA fragments obtained from the samples of the fermentation process (24, 48 and 72 h) provided the fingerprint shown in . Twenty-two bands were resolved in the PCR-DGGE analysis, corresponding to 15 different species. Only two of the analysed bands represented uncultured bacteria; and all other bands represented LAB. The LAB diversity varied across samples collected from each factory and increased from 24 to 72 h, reaching a maximum number of species at the end of the fermentation process. Lb. salivarius, Lb. helveticus and Lb. reuteri were detected only in samples from factory C, and Weissella cibaria was detected only in samples from factory B. Ws. confusa was detected in samples from factories A and B, while Lb. agilis was detected in those from factories A and C. The other seven species (Lb. acetotolerans, Lb. delbrueckii, Lb. amylovorus, Streptococcus equinus, S. lutetiensis; Lb. oris; Lb. coleohominis) were present in samples collected at each stage of the fermentation process from all three factories. These bacteria were dominant during the whole fermentation process and played an important role during fermentation.
Figure 3. DGGE profiles representing bacterial 16S rRNA gene fragments of fermented samples at different times in three factories. Fermentation time of 24 h (A1, B1, C1); 48 h (A2, B2, C2) and 72 h (A3, B3, C3) in factory A, B, C, respectively.
1: Lactobacillus acetotolerans; 2: Lb. delbrueckii; 3: Lb. salivarius; 4: Lb. helveticus; 5: Lb. reuteri; 6: Weissella cibaria; 7: Ws. confusa; 8: Lb. fermentum; 9: Lb. agilis; 10: Lb. delbrueckii; 11: Lb. amylovorus; 12: 13, 14: Lb. delbrueckii; 15: Streptococcus equinus; 16: S. lutetiensis; 17: Lb. delbrueckii; 18: Lb. oris; 19: Lb. coleohominis; 20: uncultured bacterium; 21: Lb. delbrueckii; 22: uncultured Lactobacillus sp.
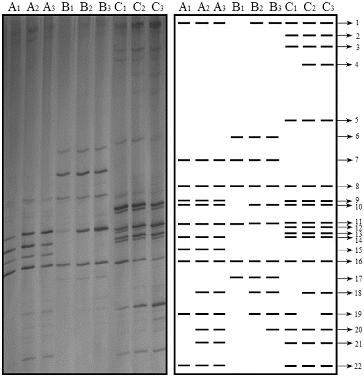
Three species were identified only by the culture-dependent method: Lb. plantarum, Lc. garvieae and E. faecalis; and seven species were detected only by the culture-independent method: Lb. acetotolerans, Ws. cibaria, Ws. confusa, Lb. agilis, S. equinus, S. lutetiensis and Lb. coleohominis. Seven species were detected by both methods: Lb. delbrueckii, Lb. salivarius, Lb. helveticus, Lb. reuteri, Lb. fermentum, Lb. amylovorus and Lb. oris. PCR-DGGE analyses confirmed the presence of the easily cultured species and also revealed the presence of several uncultured species. These two methods complement each other in the analysis of LAB diversity.
Microbial diversity can be analysed by culture-dependent and culture-independent methods, and PCR-DGGE is a useful method to identify unculturable microbial species.[Citation25] Both approaches have their advantages and disadvantages and they complement each other to give a better understanding on the microbial composition. Commonly, bacterial populations that comprise 1% or more of the total community are detectable by DGGE.[Citation26] Although the PCR-DGGE analysis could detect more microorganism species, including uncultured ones, it is recommendable to combine these two approaches for a more complete understanding of the microbial ecosystem present in the fermentation process. This is of particular importance in the rice noodle production, as commercial fermented products using mixed functional strains demonstrate better stability than traditional ones.[Citation27] More detailed analysis on the effect of the functional strains on the quality and safety of the fermented rice noodles is subject to future investigations.
Conclusions
In this study, information on the LAB ecology during the rice noodle fermentation was obtained. Lactobacillus spp. (Lb. fermentum, Lb. salivarius, Lb. plantarum), and Lactococcus spp. (Lc. garvieae) were predominant LAB throughout the fermentation process of rice noodles. These dominant species, which are potential starter cultures, might change the texture of the starch and the taste of rice noodles. However, further investigation is needed to confirm their roles. Future studies will focus on the impact of the functional strains on the quality and safety of the fermented rice noodles.
Acknowledgements
We would like to thank the rice noodle factories in Changde, Changde Administration of Quality and Technology Supervision and Changde Science and Technology Bureau for their assistance in sampling and providing the laboratory.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Liu YL. Rice products processing technology [ in Chinese]. Beijing: China Light Industry Press; 2010.
- Fu XR. Rice products processing technology and ingredient [ in Chinese]. Beijing: Chemical Industry Press; 2008.
- Lu ZH, Li LT, Cao W, Li ZG, Tatsumi E. Influence of natural fermentation on physico-chemical characteristics of rice noodles. Int J Food Sci Technol. 2003;38:505–510.
- Uchimura T, Takao T, Kikuchi K, Niimura Y, Okada S, Ohara N, Daengsubha W, Kozaki M. Identification of lacric acid bacteria isolated from fermented rice noodle Khanom jeen of Thailand. J Jpn Soc Food Sci Technol Nippon Shokuhin Kagaku Kogaku Kaishi. 1991;38:465–475.
- Nagano H, Shoji Z, Tamura A, Kato M, Omori M, To KA, Dang TT, Le VN. Some characteristics of rice paper of Vietnamese traditional food (Vietnamese Spring Rolls). Food Sci Technol Res. 2000;6:102–105.
- Ikeda M, Katoh M, Nagano H, Akuzawa S, Omori M. Characterization of the composition and bacteria in “Mohingar” fermented rice noodle from Myanmar. J Home Econ Japan. 2003;54:263–270.
- Lu ZH, Peng HH, Cao W, Tatsumi E, Li LT. Isolation, characterization and identification of lactic acid bacteria and yeasts from sour Mifen, a traditional fermented rice noodle from China. J Appl Microbiol. 2008;105:893–903.
- Nout MJR. Rich nutrition from the poorest – cereal fermentations in Africa and Asia. Food Microbiol. 2009;26:685–692.
- Lv XC, Huang XL, Zhang W, Rao PF, Ni L. Yeast diversity of traditional alcohol fermentation starters for Hong Qu glutinous rice wine brewing, revealed by culture-dependent and culture-independent methods. Food Control. 2013;34:183–190.
- Kittelmann S, Janssen PH. Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. FEMS Microbiol Ecol. 2011;75:468–481.
- Zheng XW, Yan Z, Han BZ, Zwietering MH, Samson RA, Boekhout T, Robert Nout MJ. Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 2012;31:293–300.
- Wang HY, Zhang XJ, Zhao LP, Xu Y. Analysis and comparison of the bacterial community in fermented grains during the fermentation for two different styles of Chinese liquor. J Ind Microbiol Biotechnol. 2008;35:603–609.
- Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol. 2007;114:168–186.
- Zheng Y, Zheng XW, Han BZ, Han JS, Nout MR, Chen JY. Monitoring the ecology of Bacillus during Daqu incubation, a fermentation starter, using culture-dependent and culture-independent methods. J Microbiol Biotechnol. 2013;23:614–622.
- Adams M, Mitchell R. Fermentation and pathogen control: a risk assessment approach. Int J Food Microbiol. 2002;79:75–83.
- Van Winsen RL, Lipman LJA, Biesterveld S, Urlings BAP, Snijders JMA, Van Knapen F. Mechanism of Salmonella reduction in fermented pig feed. J Sci Food Agric. 2001;81:342–346.
- Liu H, Zhou M, Xiong L , Gao X, Zhang HX. Screening of dominant lactic acid bacteria and fermentation performance from Kunming physalis alkekengi rice noodle. J Chin Cereals Oils Assoc. 2011;26:1–6.
- Haruta S, Ueno S, Egawa I, Hashiguchi K, Fujii A, Nagano M, Ishii M, Igarashi Y. Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. Int J Food Microbiol. 2006;109:79–87.
- Kim HR, Lee AR, Kim JH, Ahn BH. Microbial dynamics of commercial Makgeolli depending on the storage temperature. J Microbiol Biotechnol. 2012;22:1101–1106.
- Todorov SD, Holzapfel WH. 6 - Traditional cereal fermented foods as sources of functional microorganisms. In: Holzapfel W, editor. Advances in fermented foods and beverages. Vol. 265. Cambridge: Woodhead Publishing; 2015. p. 123–153.
- Kunene NF, Geornaras I, von Holy A, Hastings JW. Characterization and determination of origin of lactic acid bacteria from a sorghum-based fermented weaning food by analysis of soluble proteins and amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 2000;66:1084–1092.
- Oyewole OB, Odunfa SA. Characterization and distribution of lactic acid bacteria in cassava fermentation during Fufu production. J Appl Bacteriol. 1990;68:145–152.
- Giraud E, Champailler A, Raimbault M. Degradation of raw starch by a wild amylolytic strain of Lactobacillus plantarum. Appl Environ Microbiol. 1994;60:4319–4323.
- Lu ZH, Cao W, Peng HH, Wang F, Tatsumi E, Kohyama K, Li LT. Effect of fermentation metabolites on rheological and sensory properties of fermented rice noodles. J Sci Food Agric. 2008;88:2134–2141.
- Iacumin L, Cecchini F, Manzano M, Osualdini M, Boscolo D, Orlic S, Comi G. Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol. 2009;26:128–135.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703.
- Gao XZ, Liu H, Yi XX, Liu YQ, Wang XD, Xu WS, Tong QG, Cui ZJ. Microbial floral dynamics of Chinese traditional soybean paste (Doujiang) and commercial soybean paste. J Microbiol Biotechnol. 2013;23:1717–1725.