?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Genetic diversity fosters the quintessence of speciation and species acclimatization of plants in their in situ environment. Members of Cucurbitaceae, an edible and economically vital crop family, have spread across the world, dominating the tropical regions. Thus, a study of the genetic relationships among cucurbit cultivars would throw light onto the extent of diversification among these vegetal crops. The present study endeavours to understand the phylogenetic patterns and relatedness among selected species of cucurbits, using inter simple sequence repeat (ISSR) markers, which are quick, reliable and produce sufficient polymorphisms for large-scale DNA fingerprinting purposes. A total of 117 bands, of which 57 were polymorphic, were amplified by five primers. The phylogram generated on the basis of Jacquards' similarity coefficient revealed a close genetic relationship between C. maderaspatanus and C. melo, while C. sativus, a member of the same genus, was placed as a distant relative from both species, thereby demonstrating remarkable diversification among members of the same genus.
Introduction
Advanced molecular analytical techniques amalgamate the phylogenetic studies and methods of crop breeding for conservation.[Citation1] These techniques surmount the drawbacks of conventional morphological and biochemical markers used for taxonomic identification.[Citation1,Citation2] Polymorphic markers based on short DNA sequences reveal genome variations among expressed and non-expressed regions and are quick, reliable and reproducible.[Citation3] Among the different molecular markers, inter simple sequence repeat (ISSR) markers are rapid, cost effective and do not require any sequence information of the genome under study [Citation4] or any radioactive labelling based assay.[Citation5] ISSR-polymerase chain reaction (PCR) analysis involves gene amplification of a region between two inversely oriented microsatellites placed at an amplifiable distance.[Citation2] ISSR markers have been used to resolve polymorphisms among plant accessions by generating a large number of markers that target multiple microsatellite loci distributed across the genome [Citation6] and also among highly related species.[Citation7]
Cucurbitaceae, a large vegetable crop family, comprises 960 species and 125 genera, with most of its species exhibiting valuable medicinal and ornamental properties, such as anti-diabetic, anti-leptic, anti-leukaemic potential, etc.[Citation8–11] Cucurbits, also known as the family of melons, squashes, gourds, cucumber and Luffa sp., are wild natives of Africa, Madagascar and have been propagated across various parts of the world like Asia, South and Central America and Southeast Asia.[Citation8,Citation12] Most of the plants in this family are annual vines. Divergence among several members of Cucurbitaceae within a species has been reported in several studies using different molecular genetic markers.[Citation1,Citation7,Citation13] However, very few reports are available for diversity at an inter-generic level.
In the present study, multivariate analysis of eight genomes among cucurbits was performed using five ISSR markers, to assess the magnitude of divergence among the heterotic groups and evaluate inter-generic conserved/polymorphic loci. The present study also aimed to lay the foundation for the development of strategies for genetic analysis and crop improvement of these vegetal species.
Materials and methods
Plant material and DNA extraction
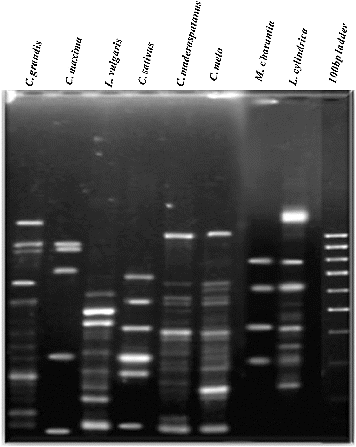
Seeds of eight cucurbit members, Cucurbita maxima Duchesne ex Lam., Lagenaria vulgaris Ser., Coccinia grandis, Cucumis sativus, Cucumis maderaspatanus, Cucumis melo, Momordica charantia and Luffa cylindrica, were collected from VNR Seeds Pvt. Ltd (India). The plants were cultivated and maintained in the green house at the School of Biotechnology and Bioinformatics, D.Y. Patil University, Navi Mumbai, under standard agronomic conditions. The samples were thoroughly rinsed with tap water and 70% alcohol and blot dried prior to use.
The DNA from all the samples was extracted by the modified cetyltrimethylammonium bromide (mCTAB) method described by Mala et al. [Citation14] and was quantified using an ultraviolet (UV)-visible spectrophomoter (Pharmaspec, UV-visible 1700, Shimadzu). The purity of the extracted DNA was estimated by the ratio of A260/A280 and by resolving the DNA in 1% agarose gel with ethidium bromide (analysed on a GeneSnap gel documentation system, SynGene).
ISSR-PCR analysis
Genomic DNA (100 ng/μL) was subjected to PCR amplification using 20 ISSR primers (15–23-mer oligonucleotides) selected and synthesized from Eurofins MWG Operon LLC (USA), based on previous studies.[Citation10,Citation13] Of these, five ISSR markers demonstrating reproducible and highly polymorphic loci were used for analysis of diversity among all plant samples selected for the study. The standard 12.5 µL PCR reaction mix comprised 100 ng of template DNA, 1X assay buffer, 1 mmol/L deoxyribonucleoside triphosphates (dNTPs), 1/2 mmol/L MgCl2 and 1 U Taq DNA polymerase, and the amplification was performed using a thermal cycler (Eppendorf, Mastercycler Gradient) with one cycle of 5 min at 94 °C, 35 cycles of 40 s at 94 °C, 40 s of annealing temperature (), 1 min at 72 °C and a final extension of 8 min at 72 °C. The amplicons were separated in a 2% agarose gel and were analysed using the GeneSnap gel documentation system (SynGene).
Table 1. Amplified fragments of eight cultivars using ISSR-PCR analysis.
Data analysis
Patterns of the amplicons were scored in a binary matrix, where the presence of reproducible polymorphic band was marked as 1 (present) or 0 (absent) and each character state was treated independently. Only consistent, bright, reproducible bands were considered for analysis. Clearly detectable amplified products ranged from 200 to 1600 bp in size. The polymorphic information content (PIC) and resolving power of each primer (Rp) were calculated for each primer as follows [5]:
where L is the total number of loci and pi is the frequency of the ith allele at the locus.
where Ibi describes the relative band informativeness and is calculated as Ibi = 1 − [2 × |0.5 − pi|], pi is the proportion of the accessions containing the ith band.
The data obtained by scoring the ISSR profiles with all the primers individually as well as collectively were subjected to similarity matrix construction using the Jaccard's similarity coefficient (Jc), using the formula [10]
where Na is the number of amplified fragments in sample A, Nb is the number of amplified fragments in sample B and Nc is the number of bands shared by samples A and B.
The neighbour-joining clustering was done on the basis of Jaccard's similarity coefficient calculated based on the binary data using the SPSS (14.0) software.
Results and discussion
Conventional methods of classification segregate plants on the basis of their morphological and anatomical traits along with a gamut of biochemical constituents. However, these factors are dependent on extraneous geo-edaphic factors and, hence, may not be reproducible for all sub-types/varieties.[Citation15,Citation16] Biodiversity studies based on genetic traits offer a rapid, reliable and reproducible tool for multivariate analysis among plants.[Citation17] In the present study, the genetic diversity and phylogenetic relationship among eight members of Cucurbitaceae were assessed using ISSR markers, which corresponds to microsatellite repeats in the genome. The five primers used herein were very effective in differentiating the genotypes.
Genetic variability based on ISSR analysis
Out of 117 reproducible amplicons generated by five ISSR primers, 57 were polymorphic. The size of the amplified products varied from approximately 100 to 1400 bp. The number of amplicons per primer ranged from 11 (I-4) to 37 (I-11), with an average of 23 amplicons per primer. The average number of polymorphic amplicons per primer was 11.4. The percentage of polymorphism ranged from 23% (I-12) to 59% (I-11), with an average of 41.71% (). The maximum number of polymorphic amplicons (22) was obtained with the I-11 primers (). The average PIC value was 0.84 and ranged from 0.23 (I-15) to 0.98 (I-11). The I-11 and I-12 primers had the highest PIC values. The resolving power ranged from 5.75 (I-4) to 33.25 (I-11) with an average of 31.3. The Jaccard's similarity coefficients ranged from 0.06 to 0.73, with an average of 0.45 ( and ). Thus, the five primers used herein were very effective in differentiating the genotypes. Approximately 35–40 monomorphic loci were observed, to the best of our knowledge, revealing for the first time putative conserved loci among the studied genera of Cucurbitaceae.
Table 2. Jaccard's similarity coefficient among cucurbit members determined on the basis of amplification with primer I-11.
Deeper insight into the assortment of genes and genetic differentiation could thus endow researchers with a foundation for developing inter/intra-genus breeding strategies.[Citation18] It could also give information about the degree of heterosis within a family in response to acclimatization drift.[Citation19] An understanding of the divergence among cultivars at an intra-generic level would also provide a valuable tool for dating the molecular evolutionary clock and revealing the ancestral pattern among these vegetal crop species. Moreover, the studied simple sequence repeats can be used as probes for mapping agronomically important traits among these species and for developing linkage maps.[Citation20]
Cluster analysis based on ISSR profiles
A collective phylogram generated from the binary matrix characters is presented in . The genotypes were grouped in five clusters at Node I (L. vulgaris) and Node V (C. maderaspatanus). The dendogram also revealed the relative magnitude of resemblance among different clusters. All the indigenous genotypes were grouped in one cluster (Node V), except C. sativus, which was placed at Node I. Also the bitter gourd (Momordica charantia) shared the node of genus Cucumis, thus revealing a common ancestral relation among them. Consequently, cucumber (Cucumis sativus), madras cucumber (Cucumis maderaspatanus) and musk melon (Cucumis melo), which are considered to be in the same genera, demonstrated a vast intra-generic variation ( and ). The cluster analysis based on Jaccards similiarity coefficient revealed very close ancestral genetic relatedness among two species of Cucumis (C. melo and C. maderaspatanus) and M. charantia, with an average similarity coefficient of 0.45, although they both belong to different sub-families and genera. Moreover, these two genera demonstrated only 22.5% similarity with C. sativus, with a similarity coefficient of 0.25 (). However, previous studies by Zhang et al. [Citation17] and Dey et al. [Citation21] report a very high genetic similarity between the C. sativus and C. melo varieties based on molecular and morphological data. The studies of Esmailnia et al. [Citation22] on 30 Iranian cucurbits, using 11 ISSR primers, and of Mohammed et al. [Citation23] on 10 cucurbits, using 13 Random Amplified Polymorphic DNA (RAPD) primers, also revealed a genetic similiarity in the range of 0.1–0.67. The intra-generic variance in our analysis can be attributed to the microsatellite repeat pattern and evolutionary diminutive evolutionary adaptations of the genotypes under study.[Citation24,Citation25] Evaluation of the chromosomal colinearity between C. sativus, C. melo (melon) and Citrullus lanatus (watermelon) has revealed that 5 of the 7 chromosomes of C. sativus arose by fusions of 10 ancestral chromosomes after the split between C. sativus and C. melo,[Citation1] thereby revealing the occurrence of divergence between two genomes within the same genus. The observed genetic relatedness thus calls for systematic analysis of these two genera, combining classical morpho-taxonomical/biochemical markers along with DNA markers. Moreover, the monomorphic loci presented herein can be further evaluated to study quantitative traits and conserved protein domains among these landraces for advanced molecular analysis.
Figure 2. Phylogram of the studied cucurbits based on combined ISSR primers (Jacquards' similarity coefficient; SPSS v. 14.0).
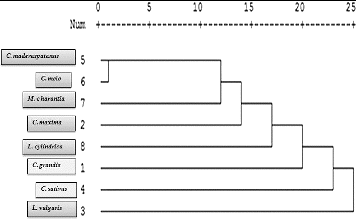
Another interesting feature observed was that members of the gourd family, viz. Coccinia grandis, clustered with C. sativus, while L. cylindrica was seen to cluster with C. maxima. Previous studies by Esmailnia et al. [Citation22] and Mohammed et al. [Citation23] have indicated C. grandis to be distantly related to C. melo and C. sativus, whereas Luffa spp. to be closely associated with C. sativus and C. melo Thus, further studies in this direction would be needed to reveal the possible clustering of these genera.
Conclusions
Based on DNA profiling using ISSR-PCR analysis, the study demonstrated genus-specific bands, which were utilized for defining the uniqueness and genetic relatedness among Cucurbitaceae members. An understanding of divergence among cultivars at intra-generic level would thus provide a valuable tool for dating the molecular evolutionary clock and ancestral pattern among vegetal crop species.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Sebastian P, Schaefer H, Telford IRH, et al. Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci U S A. 2010;107(32):14269–14273.
- Lakshmanan V, Venkataramareddy S, Neelwarne B. Molecular analysis of genetic stability in long-term micropropagated shoots of banana using RAPD and ISSR markers 2007. Electron J Biotechnol. 2007;10(1):106–113. Available from: http://www.ejbiotechnology.info/index.php/ejbiotechnology/article/viewFile/v10n1-12/205
- Pérez de la Torre M, García M, Ruth HR, et al. Analysis of genetic variability by ISSR markers in Calibrachoa caesia. Electron J Biotechnol. 2012;15(5). Available from: http://dx.doi.org/10.2225/vol15-issue5-fulltext-8
- Godwin I, Aietken E, Smith L. Application of inter simple sequence repeats (ISSR) markers to plant genetics. Electrophoresis. 1997;18(9):1524–1528.
- Saini M, Singh S, Hussain Z, et al. RAPD analysis in mungbean [Vigna radiata (L.) Wilczek.] II: a comparison of efficiency parameters of RAPD primers. Indian J Biotechnol. 2010;9:276–282.
- Sica M, Gamba G, Moniteri S, et al. ISSR markers show differentiation among Italian population of Asparagus acutifolius L. BMC Genet. 2005;6:17. Available from: http://www.biomedcentral.com/1471-2156/6/17
- Behera TK, Singh AK, Staub JE. Comparative analysis of genetic diversity in Indian bitter gourd (Momordica charantia L.) using RAPD and ISSR markers for developing crop improvement strategies. Scientia Hortic. 2008;115:209–217.
- Schaefer AH., Renner SS. Taxon. Phylogenetic relationships in the order Cucurbitales and a new classification of the gourd family (Cucurbitaceae). Taxon. 2011;60(1):122–138.
- Soundararajan R, Prabha P, Rai U, et al. Antileukemic potential of Momordica cahrantia seed extracts on human myeloid leukemia HL60 cells. Evid Based Complement Alternat Med. 2012;2012:732404. Available from: http://dx.doi.org/10.1155/2012/732404
- Singh AK, Behera TK, Chandel D, et al. Assessing genetic relationships among bitter gourd (Momordica charantia L.) accessions using inter simple sequence repeat (ISSR) markers. J Hort Sci Biotechnol. 2007;82:217–222.
- Yadav G, Mishra A, Tiwari A. Medicinal properties of ivy gourd (Cephalandra indica): a review. Int J Pharma Res Dev. 2010;2(9):92–98. Available from: http:/www.jpds.co.in/download/new/Vol.%205%20(4).pdf
- Reddy U. Cladistic analysis of a few members of Cucurbitaceae using rbcl nucleotide and amino acid sequences. Int J Bioinform Res. 2009;1(2):58–64.
- Santos MHS, Rodrigues R, Gonçalves LSA, et al. Agrobiodiversity in Cucurbita spp. Landraces collected in Rio de Janeiro assessed by molecular markers. Crop Breed Appl Biotechnol. 2012;12:96–103.
- Mala P, Parvathi JR, Payel D, et al. mCTAB: a rapid plant genomic DNA extraction method. Bionanofrontier. 2014;7(12):19–24.
- Ghazalli MN, Yunus MF, Mohammad AL. Assessment of genetic relationships of Bouea (Anacardiaceae) accessions in Peninsular Malaysia using Inter simple sequence repeat (ISSR) markers. Afr J Biotechnol. 2015;14(2):76–85.
- Dalamu, Behera TK, Gaikwad AB, et al. Morphological and molecular analyses define the genetic diversity of Asian bitter gourd (Momordica charantia L.). Aust J Crop Sci. 2012;6(2):261–267.
- Zhang C, Pratap AS, Natarajan S, et al. Evaluation of morphological and molecular diversity among Asian germplasms of Cucumis sativus and Cucumis melo. ISRN Agron. 2012;2012:134134. Available from: http://dx.doi.org/10.5402/2012/134134
- Bing Y, Xiaoli H, Zhenmin B, et al. Genetic variation in two sea cucumber (Apostichopus japonicus) stocks revealed by ISSR markers. Chin J Oceanol Limnol. 2007;25(1):91–96.
- Zhou Y, Zhou C, Yao H, et al. Application of ISSR markers in detection of genetic variation of among Chinese yam (Dioscorea opposita Thunb) cultivars. Life Sci J. 2008;5(4):6–12.
- Gonzalo MJ, Oliver M, Garcia-Mas J, et al. Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet. 2005;110(5):802–811.
- Dey SS, Behera TK, Anand P, et al. Correlation and path coefficient analysis in bitter gourd (Momordica charantia L.). Veg Sci. 2005;32(2):173–176.
- Esmailnia E, Arefrad M, Shabani S, et al. Genetic diversity and phylogenetic relationship of Iranian indigenous cucurbits investigated by inter simple sequence repeat (ISSR) markers. Biharean Biologist. 2015;9(1):47–54. Available from: http://biozoojournals.ro/bihbiol/cont/acc/bb_141131_Arefrad_acc.pdf
- Mohammed IA, Gumaa AGN, Kamal NM, et al. Genetic diversity among some Cucurbits species determined by random amplified polymorphic DNA RAPD marker. Int J Plant Res. 2012;2(4):131–137.
- Djè Y, Tahi CG, Zoro AIBi, et al. Use of ISSR markers to assess genetic diversity of African edible seeded Citrullus lanatus landraces. Scientia Hortic. 2010;124(2010):159–164.
- Maggs G-Kolling, Madesen S, Chritiansen JL. Genetic marker techniques in the family Cucurbitaceae. Genet Resour Crop Evol. 2000;47:385–393.