ABSTRACT
Rapid and inexpensive preparation of genomic DNA from rice seeds for marker-assisted selection and seed purity estimation is a major bottleneck for plant breeders. Here, we describe a high-throughput method that provides DNA at sufficient quantity and quality for these applications. Optimization of buffer composition and individual protocol stages allow processing of 384 samples within 2 h, yielding templates that reliably support downstream polymerase chain reaction of single copy amplicons up to 1.2 kb.
KEYWORDS:
Introduction
Rice is the most important staple food crop for more than half of the world's population. To adapt to the world's increasing demand for rice, several approaches are carried out to increase rice yields, including marker-assisted selection (MAS) and hybrid rice production. DNA isolation is a basic and key step for such procedures [Citation1,Citation2] and isolating DNA from seeds has at least two main advantages. First, part of the endosperm is sufficient for DNA isolation, leaving the corresponding viable embryo available for plant growth and phenotyping, which is important in MAS programmes. Second, isolating DNA from seeds accelerates genotyping by eliminating plant germination and growth, which can be very valuable in cases of seed purity identification before the sowing season.
Compared to leaf-based DNA isolation methods,[Citation3–5] seed-based DNA isolation methods in rice are rather limited.[Citation6,Citation7] The presence of stored seed reserves, including carbohydrates, lipids, proteins and polyphenols, complicates greatly the process of isolating DNA from seeds. High-quality plant DNA extraction methods tend to be time-consuming, laborious and expensive because of the multiple manipulation steps, enzymes and chemicals involved.[Citation6,Citation8] Conversely, rapid rice seed extraction methods yield DNA of poor quality that is typically insufficient for generating polymerase chain reaction (PCR) products larger than 500 bp.[Citation7,Citation9–12] In practice, PCR markers for MAS or seed purity identification are often 1 kb or more.[Citation13–15] Commercial kits (e.g. Qiagen DNeasy 96 Plant Kit) are available and effective but expensive, while automated DNA extraction system are also effective but carry high initial equipment cost and high expenses per assay which typically exceed the budgets of plant molecular breeding laboratories in developing countries.
To address this problem, here we describe a simple, rapid, high-throughput and low-cost method to prepare genomic DNA from rice seeds. The method is suitable for MAS programmes and quick identification of seed purity in practice.
Materials and methods
Materials
Rice seeds of Nipponbare (japonica) and Junyou522 (indica) were obtained from Hubei Provincial Key Laboratory for Protection and Application of Special Plants in Wuling Area of China, Key Laboratory of State Ethnic Affairs Commission for Biological Technology, College of Life Science, South-Central University, China. rTaq DNA polymerase and deoxynucleoside triphosphate (dNTP) mixture (2.5 mM) were purchased from TAKARA Biotechnology Co., LTD (Dalian). Primer synthesis was carried out by Beijing Liuhe Huada Genomics Technology Co., Ltd. α-Amylase was purchased from Worthington Biochemical Corporation. All other reagents were of analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd.
Reagents and consumables
The following reagents and consumables were used: CTAB (hexadecyl trimethyl ammonium bromide); chloroform; isopropanol; ethanol; 3 mm steel beads; 96 round well blocks with silicone cover (supplied by Geno); and 96-well PCR plates.
Solutions
The following solutions were prepared:
Extraction buffer: 200 mmol/L Tris-Cl (pH = 8.0), 50 mol/L ethylenediaminetetraacetic acid (EDTA; pH = 8.0), 2.8 mol/L NaCl, 4% CTAB, 2% sodium dodecyl sulphate;
5 mol/L potassium acetate isopropanol;
α-amylase (15 mg/mL);
Tris-EDTA buffer.
Equipment
The equipment included: Grinder (SPEX SamplePrep 2010, Geno, USA); multichannel pipettes (Eppendorf, Germany); plate mixer; plate centrifuge (Herseus Multifuge X1R, Thermo Fisher, Germany); water bath or oven or heating block, any type that can maintain temperature at 65 °C; NanoDrop ND-1000 Spectrophotometer (NanoDrop Technology, USA).
Protocol
The DNA extraction protocol includes the following stages:
Cut the rice seeds into two halves (if necessary), grind the embryo-free portion in the 96 round well block (two steel beads in each well) twice for 4 min in Geno grinder.
Add 150 µL of extract solution and 20 µL of α-amylase (15 mg/mL) to each well, keep the plates at 65 °C for 30 min.
Centrifuge plate at 1660 g for 10 min at room temperature.
Add 20 µL of chloroform and mix for 10 s by plate mixer.
Centrifuge plate at 1660 g for 1 s at room temperature, then add 20 µL of 5 mol/L potassium acetate and mix for 10 s.
Centrifuge plate at 1660 g for 10 min at room temperature.
Carefully transfer 100 µL of supernatant to a 96-well PCR plate.
Add 80 µL of isopropanol and mix by inversion several times.
Centrifuge plate at 3000 g for 15 min at room temperature.
Discard the supernatant and wash the pellet with 70% ethanol (v/v).
Dry the pellet by centrifugation at 17 g for 10 s at room temperature.
Dissolve in 50 µL of Milli Q water and directly use as template for PCR amplification.
PCR analysis
PCR analysis was performed in 10 µL of reaction mixture. A reaction tube contained 1.0 µL of template DNA, 0.2 U of rTaq, 100 µmol/L of each dNTP, 1× TAKARA rTaq buffer, and 0.5 µmol/L primers (). Amplifications were performed in the PTC-100TM PCR system (MJ, USA) with touchdown PCR with the following conditions: 94 °C for 3 min; 15 cycles at 95 °C for 30 s, annealing temperature decrement of 0.7 °C every cycle (from 65 °C to 54.5 °C) and 72 °C for 1.5 min; 20 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1.5 min; and a final extension at 72 °C for 10 min.
Table 1. PCR primers and their amplification result.
Amplification products were separated in a 1% agarose gel, stained with ethidium bromide and visualized on TFL-40 gel documentation system (Synoptics Ltd, UK).
Results and discussion
Total DNA isolated from embryo-free halves of rice seeds was checked by a NanoDrop ND-1000 Spectrophotometer. Our procedure yielded 3–7 ng/µL DNA. The ratio of absorbance at 260 nm to 280 nm was 1.8–2.1, which indicated insignificant levels of contaminating proteins and polysaccharides but high RNA content without RNase A treatment. Our method significantly minimizes the time and the use of laboratory materials. A single person can process as many as 384 samples within 2 h.
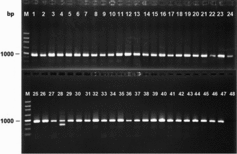
To test the suitability of the purified DNA for PCR-based analysis, several single-copy gene targets were amplified (). As shown in , the PCR amplifications were all successful, using specific primers JU-1 to identify the seed purity of hybrid rice JY522. Only one out of 47 amplicons yielded two bands, showing that seed purity for the sample was 98%. PCR amplifications were all successful when PCR the product size was less than 1.2 kb, and the success rate was 50% for PCR product sizes above 1.6 kb ().
Figure 1. Agarose gel electrophoretogram of rice seed DNA amplified by PCR using JU-1 reverse and forward primers.
α-Amylase is a critical component of our methodology, as it significantly improves the quality of the isolated rice DNA. Contamination with high concentrations of polysaccharide, which is an inhibitor for the enzymes in the case of downstream processes such as PCR, is a big problem with many seed DNA extraction methods. α-Amylase has proven to be efficacious and is used commercially to remove polysaccharide contamination and increase PCR amplification rates in DNA extraction of corn, wheat and potato (α-Amylase Ultrapure, Nippon Gene, Japan).[Citation16] In our study, PCR amplification using the primer pair Pibdom () was very unstable using DNA isolated by the same method but without the application of α-amylase (data not shown). Applying an appropriate concentration of α-amylase in the extraction buffer resulted in the purification of DNA that supported PCR amplification using primer Pibdom in all cases.[Citation17–19] Furthermore, the success rate of PCR amplification could be significantly increased for amplicons larger than 1 kb (data not shown). This indicates that our method could be applied to the DNA extraction of other cereal grains.
As mentioned above, the DNA quality of rapid isolation methods from rice seeds is typically insufficient to support PCR amplifications greater than 500 bp.[Citation20,Citation21] Even using a commercial kit (QuickExtract™ Seed DNA Extraction Solution, Epicentre, Madison, WI), the reproducibly successful PCR product sizes are still less than 750 bp.[Citation22] This could be due to DNA degradation and more contamination in these methods. PCR amplifications of DNA samples obtained by our method all succeeded for PCR product sizes up to 1.2 kb, which has high practical value in MAS programmes and quick identification of rice seed purity.
Conclusions
Overall, the method described here provides an efficient way for rapid isolation of DNA from rice seeds. The method is simple, reasonably high-throughput in nature, cost competitive compared to many other rice DNA extraction methods and, importantly, workable in laboratories lacking specialized or sophisticated instrumentation. Time and resources are saved by our method for at least two reasons: (1) the time needed for seed germination is eliminated; and (2) greenhouse and/or field space is saved. For molecular breeding applications such as MAS, positional cloning, allele mining or seed purity identification, we believe our method could be immensely helpful in DNA extraction from a large number of rice seeds.
Acknowledgments
We gratefully acknowledge Prof. Andy Flavell (University of Dundee) for critical reading of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Paris M, Carter M. Cereal DNA: a rapid high-throughput extraction method for marker assisted selection. Plant Mol Biol Rep. 2000;18:357–360.
- Mace E, Buhariwalla K, Buhariwalla H, et al. A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol Biol Rep. 2003;21:459–460.
- Sun C, He YH, Chen G, et al. A simple method for preparation of rice genomic DNA. Rice Sci. 2010;17:326–329.
- Xin ZG, Chen JP. A high throughput DNA extraction method with high yield and quality. Plant Methods. 2012;8:26.
- Xu X, Kawasaki S, Fujimura T, et al. A protocol for high-throughput extraction of DNA from rice leaves. Plant Mol Biol Rep. 2005;23:291–295.
- Mutou C, Tanaka K, Ishikawa R. DNA extraction from rice endosperm (including a protocol for extraction of DNA from ancient seed samples). In: Henry RJ, Furtado A, editors. Cereal genomics: methods and protocols. Vol. 1099, Methods in molecular biology. New York: Humana Press; 2014. p. 7–15.
- Komori T, Nitta N. A simple method to control the seed purity of japonica hybrid rice varieties using PCR-based markers. Plant Breed. 2004;123:549–553.
- Ren X, Zhu X, Warndorff M, et al. DNA extraction and fingerprinting of commercial rice cereal products. Food Res Int. 2006;39:433–439.
- Collard BCY, Das A, Virk PS, et al. Evaluation of ‘quick and dirty’ DNA extraction methods for marker-assisted selection in rice (Oryza sativa L.). Plant Breed. 2007;126:47–50.
- Fitzgerald FK, Burden DW. Evaluation of the Synergy™ rapid plant DNA isolation chemistry. Random Primers. 2014;13:1–7.
- Singh VK, Singh VK, Ellur RK, et al. Validation of rapid DNA extraction protocol and their effectiveness in marker assisted selection in crop plants. Indian J Genet. 2015;75:110–113.
- Xalxo SM, Saxena RR, Pali V, et al. A rapid and low cost method of DNA isolation for marker assisted selection in rice. Agric Res. 2014;3:83–86.
- Khanna A, Sharma V, Ellur RK, et al. Development and evaluation of near-isogenic lines for major blast resistance gene(s) in Basmati rice. Theor Appl Genet. 2015;128:1243–1259.
- Li YP, Tan YP, Xu X, et al. Establishment of specific molecular markers and application for new variety Junyou 522 in ice (Oryza sativa). J Agric Biotechnol. 2013;11:1295–1301.
- Imam J, Alam S, Mandal NP, et al. Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based makers. Euphytica. 2015;196:199–211.
- Example use of α-Amylase Ultrapure [Internet]. Toyama: Nippon Gene; c2015. [ cited 2015 August 21]. Available from: http://nippongene.com/siyaku/product/extraction/a-amylase-ultrapure/a-amylase-ultrapure.html. Japanese.
- Reference material 3 of National Food Research Institute, Japan: classification of rice cultivars by DNA analysis. Available from: http://www.hinsyu.maff.go.jp/pvr/dna_manual/san03.pdf. Japanese.
- Reference material 8 of National Food Research Institute, Japan: classification of rice seeds by DNA analysis. Available from: http://www.hinsyu.maff.go.jp/pvr/dna_manual/san08.pdf. Japanese.
- Manabu I, Jun Y, Shinichi Y, et al. Verification of DNA extraction methods for genetically modified rice. Annu Rep Okayama Prefectural Res Cent Environ Public Health. 2008;32:151–155. Japanese.
- Fouladvand A, Yari H, Emami A, et al. Optimization of a rapid DNA extraction protocol in rice focusing on age of plant and EDTA concentration. J Med Bioeng. 2013;2:218–223.
- Jhala VM, Mandaliya VB, Thaker VS. Simple and efficient protocol for RNA and DNA extraction from rice (Oryza sativa L.) for downstream applications. Int Res J Biol Sci. 2015;4:62–67.
- Manufacturer protocol of QuickExtract™ Seed DNA extraction solution. Epicentre; [cited 2015 Aug 21]. Available from: http://www.epibio.com/docs/default-source/protocols/quickextract-seed-dna-extraction-solution.pdf
