ABSTRACT
Domestic guinea pig is a model animal for human disease research. Uridine diphosphate glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1) is an important human disease-related gene. In this study, the complete coding sequence of domestic guinea pig gene UGT1A1 was amplified by reverse transcription-polymerase chain reaction. The open reading frame of the domestic guinea pig UGT1A1 gene is 1602 bp in length and was found to encode a protein of 533 amino acids. Sequence analysis revealed that the UGT1A1 protein of domestic guinea pig shared high homology with the UGT1A1 proteins of degu (84%), damara mole-rat (84%), human (80%), northern white-cheeked gibbon (80%), Colobus angolensis palliatus (80%) and golden snub-nosed monkey (79%). This gene contains five exons and four introns, as revealed by the computer-assisted analysis. The results also showed that the domestic guinea pig UGT1A1 gene had a close genetic relationship with the UGT1A1 gene of degu. The prediction of transmembrane helices showed that domestic guinea pig UGT1A1 might be a transmembrane protein. Expression profile analysis indicated that the domestic guinea pig UGT1A1 gene was differentially expressed in detected domestic guinea pig tissues. Our experiment laid a primary foundation for using the domestic guinea pig as a model animal to study the UGT1A1-related human diseases.
Introduction
Uridine diphosphate (UDP) glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1) encodes UDP-glucuronosyltransferase, an enzyme of the glucuronidation pathway that transforms small lipophilic molecules, such as steroids, bilirubin, hormones and drugs, into water-soluble, excretable metabolites (http://www.ncbi.nlm.nih.gov/gene/54658). UGT1A1 is also an important gene related to some serious human diseases. Mutations in this gene result in the reduced activity of bilirubin UDP-glucuronosyltransferase and this may lead to Crigler–Najjar syndrome types I and II and Gilbert's syndrome.[Citation1,Citation2] Zhou et al. [Citation3] indicated that UGT1A1 is associated with bilirubin kinetics during the first week of life and neonatal hyperbilirubinemia in Chinese newborns. Bilirubin levels and their changes during the middle and late parts of the first week are attributed to the variants and haplotypes in UGT1A1 gene. Khan et al. [Citation4] also reported that the UGT1A1 gene mutations result in the inherited nonhemolytic unconjugated hyperbilirubinemia in Pakistani children. Dabke et al. [Citation5] showed that the bilirubin UDP-glucuronosyltransferase isozyme 1 UGT1A1 (TA)7/(TA)7 configuration is one of the factors responsible for hyperbilirubinemia in Indian beta thalassemia patients. Similarly, Žaja et al. [Citation6] revealed that UGT1A1 TATA-box polymorphism is an important risk factor for developing jaundice in term breastfed newborns, which may be presented as either an early non-physiologic hyperbilirubinemia or breast milk jaundice in breastfed newborns – an early presentation of Gilbert's syndrome.
As mentioned above, UGT1A1 is an important human disease-related gene. Domestic guinea pig is a model animal which has been successfully used for human disease research works.[Citation7,Citation8] Until today, the domestic guinea pig UGT1A1 gene has not been reported yet. In this study, we isolated the complete coding sequence of domestic guinea pig UGT1A1 gene and subsequently performed sequence and tissue expression analyses. These would establish the primary foundation of using domestic guinea pig as a model animal to study the UGT1A1-related human diseases.
Materials and methods
Samples collection, RNA extraction and first-strand cDNA synthesis
Six adult domestic guinea pigs (Cavia porcellus; three female and three male) were obtained from the markets of Kunming (a city of China) and sacrificed. Small intestine, spleen, lung, muscle, fat, liver, heart and kidney samples were collected and frozen at –80 °C. Total RNA was extracted by using the Total RNA Extraction Kit (Gibco, USA). The RNA samples were used to perform reverse transcription for the production of the first-strand cDNA.[Citation9] These first-strand cDNAs were used to perform reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time polymerase chain reaction (qRT-PCR).
Isolation of the coding sequence
The complete coding sequence of domestic guinea pig UGT1A1 gene was amplified by RT-PCR (Thermal cycler Hema 96TC, Zhuhai, China) using the produced cDNA as a template. The 25 μL reaction system contained 2.0 μL cDNA, 2.5 μL 2 mmol/L mixed dNTPs, 2.5 μL 10 × Taq DNA polymerase buffer (with Mg2+; TaKaRa, Dalian, China), 1.0 μL 10 mmol/L forward primer 1 (5'-ATG GCC ACA GCA TCC CGG-3'), 1.0 μL 10 mmol/L reverse primer 1 (5'-TCA ATG TGT CTT GGA TTT CTG-3'), 2.0 U of Taq DNA polymerase (1 U/μL) and 14 μL sterile water. The PCR programme initially started with a 94 °C denaturation for 4 min, followed by 35 cycles at 94 °C/1 min, 59 °C/1 min, 72 °C/1 min, extension at 72 °C for 10 min and a final termination of the reaction at 4 °C. The PCR product was checked on 1% agarose/EtBr gel by running electrophoresis in 1 × tris-acetate-ethylenediaminetetraacetic acid buffer (SHENGGONG, Shanghai, China) at 90 V and then sequenced bidirectionally by using the commercial fluorometric method (SHENGGONG, Shanghai, China).
Quantitative real-time PCR for tissue expression profile analysis
qRT-PCR for evaluating the level of mRNA for UGT1A1 gene was performed on the ABI Prism 7300 Sequence Detection Systems (Applied Biosystems, Foster City, CA, USA). PCR reactions for each sample were carried out in 25 µL reaction volume containing 1 µL SYBR Green real-time PCR Master Mix, 100 ng cDNA template and 200 nmol/L of each primer (). The conditions for the real-time PCR were an initial denaturation at 95 °C for 3 min, 40 cycles at 95 °C for 15 s, optimal annealing temperature (Ta) for each specific primer for 15 s (), 72 °C for 20 s. For each sample, the reactions were run in triplicate to ensure the reproducibility of the results. The gene relative expression levels were quantified relative to the expression of the reference gene, beta actin (GenBank Accession No. AF508792), by employing the 2−ΔΔCt value model.[Citation10]
Table 1. qRT-PCR primers for domestic guinea pig UGT1A1 and actin genes.
Sequence analysis
cDNA sequence prediction was conducted by using GenScan software (http://genes.mit.edu/GENSCAN.html). The protein analysis was performed by using the basic local alignment search tool (BLAST) in the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST) and the Clustalw software (http://www.ebi.ac.uk/clustalw). Phylogenetic tree was constructed by using the dendrogram procedure of ClustalW software (http://www.genome.jp/tools/clustalw/). The theoretical isoelectric point (pI) and molecular weight (Mw) of the protein were also computed using the Compute pI/Mw Tool (http://www.expasy.org/tools/pi_tool.html). The prediction of the transmembrane helices in the protein was performed by using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Data analysis
The gene relative expression levels are presented as mean values ± standard deviation. Statistical analysis was performed using SAS software version 8.1 (SAS Institute Inc., NC, USA).
Results and discussion
Isolation of domestic guinea pig UGT1A1 gene
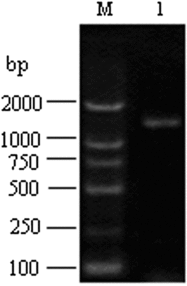
The resulting product for domestic guinea pig UGT1A1 gene, obtained through RT-PCR with pooled tissue cDNA, was found to be 1602 bp in length ().
Sequence analysis
The cDNA sequence analysis revealed that this PCR product was not homologous to any of the known domestic guinea pig genes and it was then deposited into the GenBank database (accession number: KT160222).
The sequence prediction showed that this 1602-bp cDNA sequence represented one single gene which was found to encode 533 amino acids (). The pI of the domestic guinea pig UGT1A1 protein was found to be 8.59 and the molecular weight of the putative protein was 59,688.00 Da.
Further BLAST analysis of this protein revealed high homology between the domestic guinea pig UGT1A1 protein and the UGT1A1 proteins of degu (accession number: XP_012371510, 84%), damara mole-rat (accession number: XP_010624402, 84%), human (accession number: AFN10615, 80%), northern white-cheeked gibbon (accesession number: XP_003278588, 80%), Colobus angolensis palliatus (accesession number: XP_011785026, 80%) and golden snub-nosed monkey (accesession number: XP_010366744, 79%) ().
Figure 3. Alignment of proteins encoded by the UGT1A1 gene in domestic guinea pig and in six other species.

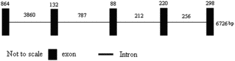
To obtain the genomic DNA of UGT1A1, the publicly available domestic guinea pig genome database at the NCBI Genome Resources (http://www.ncbi.nlm.nih.gov/projects/genome) was screened by using the full-length cDNA sequence of UGT1A1 gene as a seed by BLAST analysis. A bacterial artificial chromosome clone CH234-357O12 (GenBank accession no. AC189144.2) was found to encompass the entire UGT1A1 gene (57,303 bp–64,028 bp). The UGT1A1 gene was found to be 6726 bp in length and consisted of 5 exons and 4 introns. All exon–intron splice junction sequences conformed to the GT–AG rule ().
Figure 4. Genomic sequence organization representing the open reading frame of the domestic guinea pig UGT1A1 gene.
The transmembrane helices prediction showed that the domestic guinea pig UGT1A1 protein might be a transmembrane one ().
Based on the results of the UGT1A1 alignment of different species' proteins, a phylogenetic tree was constructed using the Clustalw software, as shown in . The phylogenetic tree analysis revealed that the domestic guinea pig UGT1A1 gene might have a close genetic relationship with that of degu.
Tissue expression profile
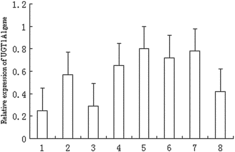
The tissue expression analysis indicated that the domestic guinea pig UGT1A1 gene was highly expressed in muscle and liver, moderately expressed in heart, spleen, kidney and small intestine and weakly expressed in fat and lung ().
Comparative genomics research has revealed that virtually all (99%) of the protein-coding genes in humans align with homologs in mouse, and over 80% are clear 1:1 orthologs for human and mouse which are both mammalian species.[Citation11] This extensive conservation in protein-coding regions implied that such conservation of protein-coding sequences may be expected in domestic guinea pigs and other mammalian animals. From the sequence analysis of UGT1A1 genes, it can be seen that the coding sequences of UGT1A1 genes were highly conserved in some mammalians, including human and domestic guinea pig. Comparative analysis of the genomic structures in different mammalian species also showed that the genomic structures of UGT1A1 genes were highly conserved in some mammalians, including human and domestic guinea pig (data not shown). They all have five exons (same length) and four introns (some with different length). This implied that these mammalian UGT1A1 genes should have similar functions. We can use the domestic guinea pig as a model animal to study the UGT1A1-related diseases of other mammalians, including human. We also noticed that the domestic guinea pig UGT1A1 protein showed 80% identity to the human UGT1A1 protein. This implied that when we use the domestic guinea pig as a model animal to study the human UGT1A1-related diseases, we should pay more attention to this difference.
Variants in the UGT1A1 gene have been reported to be associated with Crigler–Najjar syndromes types I and II and Gilbert's syndrome.[Citation1–6] In our experiments, no variations were detected among the six animals. To study the presence of variations in the UGT1A1 gene, further research should be carried out including a larger number of animals.
From the tissue distribution analysis in our experiment, it can be seen that the UGT1A1 gene was obviously expressed in all tissues. As we know, phenotypic variances are mainly determined by the gene expression differences so that it can be inferred that the UGT1A1 gene might play an important role in the biological metabolism of domestic guinea pig. This coincides with the fact that the UGT1A1 functions as an enzyme of the glucuronidation pathway. It can transform small lipophilic molecules, such as steroids, bilirubin, hormones and drugs, into excretable metabolites, which are water-soluble (http://www.ncbi.nlm.nih.gov/gene/54658). We also found that the UGT1A1 gene was differentially expressed in some tissues. The suitable explanation for this, under the present conditions, is that these biological functions of UGT1A1 gene might be presented diversely in different tissues.
Conclusions
In conclusion, we first isolated the domestic guinea pig UGT1A1 gene. We found that the domestic guinea pig UGT1A1 gene has a 1602-bp open reading frame encoding a protein which shared high homology with other 6 UGT1A1 proteins. This gene had a close genetic relationship with the UGT1A1 gene of degu. Furthermore, the domestic guinea pig UGT1A1 protein might be a transmembrane protein. The results also indicated that the domestic guinea pig UGT1A1 gene was differentially expressed in various tissues. Our experiment laid a primary foundation for using the domestic guinea pig as a model animal to study the UGT1A1-related human diseases.
Disclosure statement
No potential conflict of interests was reported by the authors.
Additional information
Funding
References
- Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616.
- Suzuki M, Hirata M, Takagi M, et al. Truncated UDP-glucuronosyltransferase (UGT) from a Crigler-Najjar syndrome type II patient colocalizes with intact UGT in the endoplasmic reticulum. J Hum Genet. 2014;59(3):158–162.
- Zhou Y, Wang SN, Li H, et al. Quantitative trait analysis of polymorphisms in two bilirubin metabolism enzymes to physiologic bilirubin levels in Chinese newborns. J Pediatr. 2014;165(6):1154–1160.
- Khan S, Irfan M, Sher G, et al. UGT1A1 gene mutations in Pakistani children suffering from inherited nonhemolytic unconjugated hyperbilirubinemias. Ann Hum Genet. 2013;77(6):482–487.
- Dabke PS, Colah RB, Ghosh KK, et al. Role of co-inherited Gilbert syndrome on hyperbilirubinemia in Indian beta thalassemia patients. Hematology. 2014;19(7):388–392.
- Žaja O, Tiljak MK, Štefanović M, et al. Correlation of UGT1A1 TATA-box polymorphism and jaundice in breastfed newborns-early presentation of Gilbert's syndrome. J Matern Fetal Neonatal Med. 2014;27(8):844–850.
- Clark S, Hall Y, Williams A. Animal models of tuberculosis: guinea pigs. Cold Spring Harbor Perspect Med. 2014;5(5):a018572.
- Schuller S, Sergeant K, Renaut J, et al. Comparative proteomic analysis of lung tissue from guinea pigswith leptospiral pulmonary haemorrhage syndrome (LPHS) reveals a decrease in abundance of host proteins involved in cytoskeletal and cellular organization. J Proteomics. 2015;122:55–72.
- Liu GY, Xiong YZ, Deng CY, et al. Comparison of gene expression patterns in longissimus dorsi of pigs between the high-parent heterosis cross combination landrace large white and the mid-parent heterosis cross combination large white Meishan. Asian Australasian J Anim Sci. 2004;17:1192–1196.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Hardison RC. Comparative genomics. PLOS Biol. 2003;1(2):e58.