ABSTRACT
In this research, two well-recognized standard grape cultivars, Cabernet Sauvignon and Merlot, together with eight historical autochthonous grapevine cultivars from Eastern Anatolia in Turkey, were genetically characterized by using 12 pairs of simple sequence repeat (SSR) primers in order to evaluate their genetic diversity and relatedness. All of the used SSR primers produced successful amplifications and revealed DNA polymorphisms, which were subsequently utilized to evaluate the genetic relatedness of the grapevine cultivars. Allele richness was implied by the identification of 69 alleles in 8 autochthonous cultivars with a mean value of 5.75 alleles per locus. The average expected heterozygosity and observed heterozygosity were found to be 0.749 and 0.739, respectively. Taking into account the generated alleles, the highest number was recorded in VVC2C3 and VVS2 loci (nine and eight alleles per locus, respectively), whereas the lowest number was recorded in VrZAG83 (three alleles per locus). Two main clusters were produced by using the unweighted pair-group method with arithmetic mean dendrogram constructed on the basis of the SSR data. Only Cabernet Sauvignon and Merlot cultivars were included in the first cluster. The second cluster involved the rest of the autochthonous cultivars. The results obtained during the study illustrated clearly that SSR markers have verified to be an effective tool for fingerprinting grapevine cultivars and carrying out grapevine biodiversity studies. The obtained data are also meaningful references for grapevine domestication.
KEYWORDS:
Introduction
Anatolia and Transcaucasia have long been regarded as likely homelands of many plant species, including both cultivated Vitis vinifera ssp. sativa and wild Vitis vinifera ssp. sylvestris.[Citation1–3] Eastern and Southern Anatolia have special position and have long been affiliated with grapevine. Previously, researchers have commonly ascribed the origin of viticulture and wine making to Anatolia.[Citation4] Anatolia has a long viticulture history and a rich tradition owing to the settlement of many populations and civilizations. This situation made Anatolia a place of choice for the production and exchange of plant material. This inheritance has led not only to the existence of a quite large grapevine germplasm, but also to the presence of homonymy and synonymy cases.[Citation3] Each grape-growing region in Turkey has its exceptional local grapevine cultivars in terms of colour, taste, shape, bunch density, etc.[Citation5,Citation6] There is also a wide variation with regards to synonymous cultivars in each region. This is why, an accurate description of these cultivars is of great significance for cultivar standardization and detection of the total cultivar number.[Citation3,Citation5]
Cultivation of both early- and late-ripening grape cultivars is possible because of the diverse ecological conditions existing within Eastern Anatolia.[Citation6] Grapes produced in this region are mostly consumed as table grapes with small amounts utilized in wine-making and in snack for food industries.[Citation6] Elazig, Erzincan and Malatya provinces of this region have a rich grape germplasm and are the major viticulture areas, followed by Van, Erzurum and Igdir provinces. The existence of wild grape populations available in the region reveals that viticulture has long been known in these places. Igdir Province is the main traditional viticulture area in this region, including eight very old and local grapevine cultivars. During the Christian era, these historical cultivars were evaluated for wine-making. Contrarily, nowadays, wine production is not practised as a result of the conservative lifestyle of the people living in this region. From the point of microclimate characteristics, Igdir Province is known to be much more different in comparison with the other provinces of Eastern Anatolia. It is hoped that the grape germplasm of this region would have economically significant adaptive characteristics that can possibly be incorporated into future grape breeding programmes.[Citation5,Citation6]
Because of the great economic significance of grapevine throughout the world, germplasm collections involve plentiful grape accessions propagated clonally, but phenotypic and genetic characterizations of the accessions are still very inadequate for crop improvement. It is well understood that genetic variability is invaluable for crop and grapevine improvement and better understanding of grapevine gene function.[Citation4–6]
Nowadays, it is widely acknowledged that DNA markers represent a momentous resource for constructing genetic maps, describing distinctive individuals and assessing genetic relatedness. Microsatellites, also known as simple sequence repeat (SSR) markers or short tandem repeats, are short repeat motifs that show high level of length polymorphism due to insertion or deletion mutations of one or more repeat types. Microsatellites are a rewarding tool for the identification and characterization of identical genotypes in a germplasm collection and in grape cultivars within the context of genetic diversity researches.[Citation6–8]
In our previous research, we studied the genetic relationships among autochthonous grapevine cultivars from Northeast Turkey.[Citation6] In this study, we widened this research by studying autochthonous grapevine cultivars from Eastern Turkey. Igdir Province, located in the eastern part of Turkey, has a great number of autochthonous grapevine cultivars used for many centuries. It is important to characterize this germplasm objectively. In this context, SSR markers were used for the first time, with the intention of investigating the relationship and genetic diversity among eight autochthonous grapevine cultivars provided from Igdir Province in Eastern Anatolia, Turkey. We expect that the report documented here would be helpful for selection and more efficient utilization of this germplasm for grape breeding purposes in future.
Materials and methods
Plant material
For the genetic characterization, leaf samples from the eight autochthonous grapevine cultivars used in this study were gathered from Igdir Province in the Eastern Anatolia region, Turkey. A total number of 10 grapevine cultivars, together with two reference cultivars, Cabernet Sauvignon and Merlot, were included in the SSR analysis. presents some considerable ampelographic traits of the autochthonous grapevine cultivars evaluated genetically in the present study.
Table 1. Basic descriptive characteristics of the eight grapevine cultivars used in this study.
DNA extraction
Genomic DNA was extracted from young leaf tissues by using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) in line with the instructions provided by the manufacturer. Subsequently, an RNAse treatment was carried out on the eluted DNA samples. Both purity and concentration of the DNA were observed on 1% (w/v) agarose gels and by using NanoDrop® ND-1000 Spectrophotometer.
SSR analysis
Twelve internationally well-known grape SSR markers (i.e. VMC2C3, VMC2H4, VrZAG62, VrZAG79, VrZAG83, VVIB01, VVMD5, VVMD24, VVMD27, VVMD28, VVMD31 and VVS2) were used in the polymerase chain reaction (PCR) studies. The PCR was conducted in a final volume of 10 μL and the reaction mixture contained 15 ng genomic DNA, 5 pmol of each primer, 0.5 mmol/L dNTP, 0.5 U GoTaq DNA Polymerase (Promega), 1.5 mmol/L MgCl2 and 2 μL 5X buffer. The forward primers were labelled with WellRED fluorescent dyes D2 (black), D3 (green) and D4 (blue) (Proligo, Paris, France).
Reactions without DNA were used as negative controls. The PCR amplification was implemented by using the Biometra® PCR System. The amplification conditions consisted of an initial denaturation step of 3 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 52 °C–56 °C and 2 min at 72 °C, with a final extension at 72 °C for 10 min. The PCR products were initially separated on a 3% (w/v) agarose gel run at 80 V for 2 h. The gel was then stained with ethidium bromide at a concentration of 10 mg/mL. A DNA ladder (100 bp) (Promega) was used for the approximate quantification of the bands. The amplification products were visualized under UV light, and the estimation of their sizes was made relatively to the DNA ladder. For further identification of polymorphisms, the PCR products were run on CEQ™ 8800 XL Capillary Genetic Analysis System (Beckman Coulter, Fullerton, CA). The genetic analyses were repeated at least twice to make certain the reproducibility of the obtained results. In the study, allele sizes were estimated for each SSR locus with the use of the Beckman CEQ Fragment Analysis software. In each run, Cabernet Sauvignon and Merlot cultivars were evaluated as reference cultivars.
Genetic analysis
The genetic analysis programme ‘IDENTITY’ 1.0 [Citation9] was employed, as suggested by Paetkau et al.,[Citation10] for the calculation of the number of alleles, expected heterozygosity (He) and observed heterozygosity (Ho) and probability of identity (PI) per locus. Genetic similarity was obtained via the programme ‘MICROSAT’ (version 1.5) [Citation11] using proportion of shared alleles, calculated with ‘ps (option 1–(ps))’, as described by Bowcock et al.[Citation12] Thereafter, the obtained results were converted to a similarity matrix, and a dendrogram was constructed with the unweighted pair-group method with arithmetic mean (UPGMA) method,[Citation13] with the support of the software NTSYS-pc (Numerical Taxonomy and Multiware Analysis System, version 2.0).[Citation14]
Results and discussion
The present study illustrates the results of SSR analysis of 10 grapevine cultivars comprising eight autochthonous grapevine cultivars from Eastern Anatolia and two reference cultivars, Cabernet Sauvignon and Merlot, in order to define the genetic diversity and relatedness among the evaluated genotypes. A total number of 69 alleles, from 3 to 9 alleles per locus, with a mean value of 5.75 alleles per locus, were genetically identified for the 8 autochthonous cultivars (). Polymorphic bands were obtained with all primers. VMC2C3 and VVS2 loci were the most polymorphic among the 12 loci and had the highest effective number of alleles (nine and eight alleles, respectively). The number of alleles per loci was decreasing gradually as follows: VMC2C3 (nine alleles) > VVS2 (eight alleles) > VVMD28 (seven alleles) > VMC2H4 = VrZAG62 = VrZAG79 = VVMD24 (six alleles) > VVMD5 = VVMD31 (five alleles) > VVIB01 = VVMD27 (four alleles) > VrZAG83 (three alleles) ().
Table 2. Descriptive statistics and genetic diversity of local grape genotypes at 12 microsatellite loci.
The study revealed that the mean He and Ho were 0.749 and 0.739, respectively, over 12 loci, related to the eight autochthonous cultivars. Among the 12 loci, the highest Ho values (1.00) were recorded in VVMD31 and VVS2 loci, implying high genetic diversity, whereas the lowest (0.50) values were observed in VMC2H4, VrZAG79 and VVMD28 loci (). Under the study, the most informative loci, with respect to the PI, were VVMD28 and VVS2, consisting of seven alleles (PI = 0.068) and eight alleles (PI = 0.089), respectively. However, the least informative loci were proved to be VrZAG83 and VVIB01, which were composed of three and four alleles, respectively (PI = 0.253 and PI = 0.281, respectively) ().
Allele sizes (bp) of 10 genotypes from 12 SSR loci are given in . In general, the most frequent alleles, in regards to all loci, were 191, 195 and 233 with similar frequent ratio (5.02%) as also described in . However, the most frequent alleles in VMC2C3, VMC2H4, VrZAG62, VrZAG79, VrZAG83, VVIB01, VVMD5, VVMD24, VVMD27, VVMD28, VVMD31 and VVS2 were 163 (25%), 204 (30%), 188 and 194 (30%), 246 (45%), 191 (50%), 292 (55%), 233 (50%), 209 (30%), 195 (40%), 243 (25%), 209 (30%) and 151 (35%), respectively ().
Table 3. Allele sizes (bp) of 12 microsatellites loci from 10 grapevine cultivars.
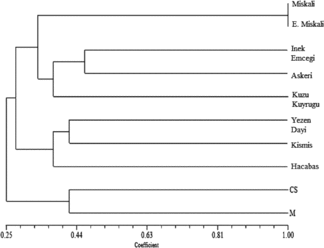
The genetic similarity was estimated within the range of 0.125–1.00 among the genotypes analysed in the study. The Kuzu Kuyrugu and Cabernet Sauvignon cultivars were the most distant with a 0.125 similarity ratio (). However, Miskali and Erkek Miskali cultivars were identical with a genetic similarity ratio of 1.00 ( and ). To clarify the genetic relationship between grapevine cultivars, a dendrogram, constructed with the results from the UPGMA cluster analysis over 12 SSR loci, classified the 10 cultivars into two main groups, as illustrated in . The international well-recognized Cabernet Sauvignon and Merlot cultivars were clustered together in cluster 1 and separated from the remaining local cultivars. The second main cluster contained eight autochthonous cultivars and this cluster was further divided into two subgroups. Three autochthonous cultivars (Hacabas, Kismis and Yezen Dayi) were included in the first subgroup. In the results of the UPGMA cluster analysis, the second subgroup involved the remaining five autochthonous cultivars (Kuzu Kuyrugu, Askeri, Inek Emcegi, Erkek Miskali and Miskali).
Table 4. Similarity ratio of local and international grape cultivars.
Figure 1. Dendrogram showing the relationship of 10 grapevine cultivars based on UPGMA cluster analysis of 12 SSR loci. Note: CS, Cabarnet Sauvignon; M, Merlot.
The present research is the first one regarding the use of SSR markers for evaluating the genetic relatedness among eight autochthonous grapevine cultivars from the eastern part of Turkey. The present results illustrated the efficient use of microsatellites in grapevine. Generally, different levels of amplifications were obtained by all the tested microsatellite primer pairs. The mean value of 5.75 alleles per locus from the used microsatellites, with the objective to fulfil polymorphic amplification patterns, is in agreement with the previously reported data of 4–16 alleles per locus in V. vinifera germplasms, evaluated by using microsatellites in several countries.[Citation15–19] The average number of alleles (5.75) in the present research was recorded in contrast with the earlier reports, which used 25 autochthonous cultivars (8.67) from northeastern Turkey,[Citation6] 33 Slovenian cultivars (8.00),[Citation20] 11 Romanian cultivars (7.90),[Citation21] 50 Greek cultivars (7.90),[Citation22] or 51 accessions from Bosnia and Herzegovina (7.82).[Citation7]
The obtained results may provide utility clues in the improvement of grapevine cultivars specific for the eastern part of Anatolia. The wide within group variation may be attributed to the introduction and spread of wild and semi-domesticated grapes, natural hybridization and human selection.
In previous studies, the highest number of alleles was recorded for VVS2 primer [Citation6,Citation16,Citation19,Citation23,Citation24] among the SSR primers. However, the lowest number of alleles in previous studies was found for VrZAG83 primer.[Citation6,Citation8,Citation19,Citation25,Citation26]
In this study, the mean Ho level was lower than that of He (0.739 and 0.749, respectively). The Ho levels were found to be in the range of 74.3%–85.5% for the grapes grown in different countries.[Citation15,Citation16,Citation18,Citation19,Citation25] In a previous study, Štajner et al. [Citation8] reported that the He varied from 0.594 at locus VChr15a to 0.889 at locus VVMD28, with an average He of 0.775 in 121 grape genotypes. The Ho was in the range from 0.315 (VChr8a) to 0.845 (VVMD32) in their study.[Citation8] High levels of heterozygosity are widely found among clonally propagated, outbreeding, perennial species including V. vinifera.[Citation27,Citation28] Grapes, as an outbreeding species, possess considerably heterozygous cultivars and are affected from severe inbreeding depression.[Citation29]
As also mentioned in the introduction section, results of SSR analysis as a utility tool for identification are highly accepted throughout the world. This means that SSR analysis permits the comparison of different grape genotypes throughout the world, in order to determine the level of heterozygosity occupied by natural and human selection for genetically assessed grape cultivars. Microsatellites (SSR fingerprinting) are a very useful tool for detection of identical genotypes in germplasm collections, as well as for detection and characterization of grape cultivars within the scope of genetic diversity investigations.[Citation30] Characterization of plant materials by using ampelographic methods may cause wrong interpretations, but SSR (DNA-based) markers, which are widely preferred in recent years, produce more trustworthy results for genetic detection [Citation31] due to the fact that the results obtained from the markers are not dependent on environmental factors. In brief, the relevance of Vitis SSR markers for genetic mapping, cultivar identification, parentage analysis, as well as genetic origin and diversity of the grape cultivars in germplasm collections, has been addressed.[Citation32]
In the literature, only bioactive compounds such as organic acids, sugars and several phenolic compounds were identified in the Igdir ancient grape cultivars.[Citation33] The present investigation is the first study to characterize genetically those ancient grape cultivars and their genetic diversity and relatedness. More data of SSR markers which will provide a noteworthy contribution to the history of the grapevine domestication need to be obtained.[Citation4] The eco-geographic adaptations must also be taken into consideration for further genetic identifications.[Citation34] All provinces in the Eastern Anatolia region and all undiscovered provinces in other regions should genetically assess grapevine cultivars with SSR markers to reveal the historical gene flows between provinces and regions.
Conclusions
The present research is the first related to the use of SSR markers for evaluating the genetic relatedness between eight autochthonous grapevine cultivars from Igdir Province of Turkey. In the research, a high genetic distance values within the range from 0.125 to 1.000 were detected by genetic evaluations of grape cultivars. The SSR data reported here might introduce worthy inputs contributing to further grapevine selection and breeding strategies for protection and exploitation of the grapevine germplasm in the region. Additionally, the present SSR data might enable researchers not only to conserve the valuable genetic resources for providing crop improvement, but also to reliably compare the SSR data from earlier and future studies on grape cultivars. The obtained data are also a worthwhile reference for grapevine domestication.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ceyhan E, Harmankaya M, Kahraman A. Combining ability and heterosis for concentration of mineral elements and protein in common bean (Phaseolus vulgaris L.). Turk J Agric For. 2014;38:581–590.
- Baloch FS, Karakoy T, Demirbas A, et al. Variation of some seed mineral contents in open pollinated faba bean (Vicia faba L.) landraces from Turkey. Turk J Agric For. 2014;38:591–602.
- Ergul A, Kazan K, Aras S, et al. AFLP analysis of genetic variation within the two economically important Anatolian grapevine (Vitis vinifera L.) varietal groups. Genome. 2006;49:467–495.
- This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006;22:511–519.
- Hizarci Y, Ercisli S, Yuksel C, et al. Genetic characterization and relatedness among autochthonous grapevine cultivars from Northeast Turkey by simple sequence repeats (SSR). J Appl Bot Food Qual. 2012;85:224–228.
- Boz Y, Bakir M, Çelikkol BP, et al. Genetic characterization of grape (Vitis vinifera L.) germplasm from Southeast Anatolia by SSR markers. Vitis. 2011;50(3):99–106.
- Tomić L, Štajner N, Jovanović-Cvetković T, et al. Identity and genetic relatedness of Bosnia and Herzegovina grapevine germplasm. Sci Hortic. 2012;143:122–126.
- Štajner N, Tomić L, Ivanišević D, et al. Microsatellite inferred genetic diversity and structure of Western Balkan grapevines (Vitis vinifera L.). Tree Genet Genomes. 2014;10:127–140.
- Wagner HW, Sefc KM. Identity 1.0. centre for applied genetics. Vienna: University of Agricultural Science; 1999.
- Paetkau D, Calvert W, Stirling I, et al. Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol. 1995;4:347–354.
- Minch E, Ruiz-Linares A, Goldstein DB, et al. Microsat (Version 1.4d): a computer program for calculating various statistics on microsatellite allele data. Stanford: Stanford University Medical Center; 1995.
- Bowcock AM, Ruiz-Linares A, Tomfohrde T, et al. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457.
- Sneathh PH, Sokal RR. Numerical taxonomy. San Francisco: Freeman; 1973.
- Rohlf FJ. NTSYS-PC numerical taxonomy and multivariate analysis system. New York: Exeter Publishing; 1988.
- Bowers JE, Dangl GS, Vignani R, et al. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome. 1996;39:628–633.
- Fatahi R, Ebadi A, Bassil N, et al. Characterisation of Iranian grapevine cultivars using microsatellite markers. Vitis. 2003;42:185–192.
- Martin JP, Borrego J, Cabello F, et al. Characterization of Spanish grapevine cultivar diversity using sequence-tagged microsatellite site markers. Genome. 2003;46:10–18.
- Martinez LE, Cavagnaro PF, Masuelli RW, et al. SSR-based assessment of Genetic diversity in South American Vitis vinifera varieties. Plant Sci. 2006;170:1036–1044.
- Tangolar SG, Soydam S, Bakir M, et al. Genetic analysis of grapevine cultivars from the Eastern Mediterranean region of Turkey, based on SSR markers. J Agric Sci. 2009;15:1–8.
- Štajner N, Rusjan D, Korošec-Koruza Z, et al. Genetic characterization of old Slovenian grapevine varieties of Vitis vinifera L. by microsatellite genotyping. Am J Enol Vitic. 2011;62(2):250–255.
- Gheorghe RN, Popescu CF, Pamfil D, et al. Genetic diversity of some Romanian grapevine cultivars as revealed by microsatellite markers. Rom Biotechnol Lett. 2010;15(2):26–31.
- Lefort F, Roubelakis-Angelakis KKA. Genetic comparison of Greek cultivars of Vitis vinifera L. by nuclear microsatellite profiling. Am J Enol Vitic. 2001;52(2):101–108.
- Nunez Y, Fresno J, Torres V, et al. Practical use of microsatellite markers to manage Vitis vinifera germplasm: molecular identification of grapevine samples collected blindly in D.O. “El Bierzo” (Spain). J Hortic Sci Biotech. 2004;79:437–440.
- Selli F, Bakir M, Inan H, et al. Simple sequence repeat-based assessment of genetic diversity in Dimrit and Germe grapevine accessions from Turkey. Vitis. 2007;46:182–187.
- Sefc KM, Lopes MS, Lefort F, et al. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor Appl Genet. 2000;100:498–505.
- Snoussi H, Slimane MHB, Ruiz-Garcia L, et al. Genetic relationship among cultivated and wild grapevine accessions from Tunisia. Genome. 2004;47:1211–1219.
- Hvarleva T, Rusanov K, Lefort F, et al. Genotyping of Bulgarian Vitis vinifera L. cultivars by microsatellite analysis. Vitis. 2004;43:27–34.
- Dzhambazova T, Tsvetkov I, Atanassov I, et al. Genetic diversity in native Bulgarian grapevine germplasm (Vitis vinifera L.) based on nuclear and chloroplast microsatellite polymorphisms. Vitis. 2009;48(3):115–121.
- Olmo HP. Grapes. In: Simmonds NW, editor. Evolution of crop plants. London: Longman; 1976. p. 294–298.
- Doulati-Baneh H, Mohammadi SA, Labra M. Genetic structure and diversity analysis in Vitis vinifera L. cultivars from Iran using SSR markers. Sci Hortic. 2013;160:29–36.
- Štajner N, Korošec-Koruza, Rusjan D, et al. Microsatellite genotyping of old Slovenian grapevine varieties (Vitis vinifera L.) of the Primorje (coastal) winegrowing region. Vitis. 2008;47(4):201–204.
- Tomić L, Štajner N, Javornik B. Chapter 1. Characterization of grapevines by the use of genetic markers. In: Poljuha D, Sladonja B, editors. The Mediterranean genetic code – grapevine and olive. Rijeka, Croatia: INTECH; 2013. p. 3–23.
- Eyduran SP, Akin M, Ercisli S, et al. Sugars, organic acids, and phenolic compounds of ancient grape cultivars (Vitis vinifera L.) from Igdir province of eastern Turkey. Biol Res. 2015;48(2):1–8.
- Zdunić G, Preece JE, Aradhya JE, et al. Genetic diversity and differentiation within and between cultivated (Vitis vinifera L. ssp. sativa) and wild (Vitis vinifera L. ssp. sylvestris) grapes. Vitis. 2013;52(1):29–32.
