ABSTRACT
Hawthorn (Crataegus spp.) is an important forest fruit species in Iran. Genetic variability among some genotypes of hawthorn was investigated using morphological traits and random amplified polymorphic DNA (RAPD) marker. The collected genotypes belonged to four species of Crataegus genus. High variability among genotypes was found for most of the traits. Results from the principal component analysis (PCA) indicated that 85.05% of the observed variability was accounted by the first five components. The first two components explained about 55.24% of the total achieved variability. In PCA, fruit weight, fruit length, fruit diameter, fruit moisture, fruit dry matter, leaf length, leaf area, leaf width, number of leaves per node, seed weight and seed length were predominant in the first two components, indicating that they were useful for the assessment of hawthorn germplasm characterization. A total of 58 polymorphic bands were produced with 10 RAPD primers. The bands' sizes ranged from 180 to 2700 bp. The number of the observed polymorphic bands for each primer ranged from 4 to 8, with an average of 5.8 alleles per locus. The polymorphism information content was observed to be the highest (0.79) in the Oligo_32 locus, whereas the Oligo_339 locus had the lowest value of 0.64, with an average of 0.72, among the RAPD primers. The Jaccard's genetic similarity coefficient ranged from 0.12 to 0.95 among the genotypes. At a similarity coefficient of 0.46, the unweighted pair group method with arithmetic mean (UPGMA) cluster analysis divided the genotypes into three major groups.
Introduction
Hawthorn (Crataegus spp.) comprises of a complex group of trees and shrubs, native to northern temperate zones, mostly between latitudes 30° and 50° N.[Citation1] Hawthorn belongs to the Maloideae subfamily in the Rosaceae family, a natural group of complex genera with the ability to interbreed freely (hybridize).[Citation1] The genus Crataegus, which has been described as the largest genus of the Maloideae subfamily, includes more than 265 hawthorn species.[Citation2] According to Christensen,[Citation3] the Crataegus species' number could vary between 150 and 1200.[Citation4] Among the 50–100 Old World species of Crataegus L., the taxa of the genus Crataegus, occurring in Europe, Northern Africa and Western Asia, stands out as having small leaves with 1–4 pairs of lobes, usually extending 0.5 times or more the width of the lamina to the midrib and ventro-laterally smooth pyrenes.[Citation3] It is assumed that a large number of phenotypically intermediate forms has resulted in introgressive hybridization between the different species and successive backcrossing with one of the parental species.[Citation5,Citation6]
A serious problem for taxonomists and evolutionary biologists is presented by hybridization, apomixes and polyploidy that frequently occurs in Crataegus. Taxonomic complexity is mostly caused by the cognition of large numbers of very narrowly defined species.[Citation7] Hybridization between two species may occur.[Citation3,Citation4] Hawthorn fruits and flowers are widely used for the treatment of congestive heart failure, because they contain flavonoids and related proanthocyanidins.[Citation8] Also, some species are grown as ornamental plants, because of their small stature, brilliant flowers during the spring and brightly coloured fruits during the fall.[Citation9] It is generally accepted that the genetic variation in plant populations is an essential prerequisite for the use of genetic resources. To date, in hawthorn, this has been based on variation of morphological traits, a method that suffers from low numbers of independent characters and often poor heritability.[Citation10] Iran is one of the genetic centres of Crataegus spp., but there are few studies that have attempted to describe the Crataegus genus in this country.[Citation11] Hawthorn, found in all regions of Iran, has a high morphological diversity, particularly among the leaf and fruit characteristics. This diversity is believed to be achieved by birds and some mammals that eat the fruits and serve as vectors in their distribution. The information of this research will be useful to identify desirable genotypes for preservation in collections and their use in hawthorn breeding programmes.
Materials and methods
Plant materials and morphological studies
Leaf and fruit samples from 30 wild hawthorn genotypes (), belonging to four species, including C. pontica (26 individuals), C. monogyna (1 individual), C. microphylla (1 individual) and C. pentagyna (2 individuals), were collected from different geographical sites in Iran, including Mazandaran, Ilam, Kordestan and Kermanshah provinces (). The fruit samples were harvested randomly from various parts of the trees at the ripening date of each genotype. Seventeen morphological variables, including length (cm), width (cm) and area (cm2) of the leaf, petiole length (cm), length of leaf lobes (cm), number of leaf lobes, number of leaves per node, fruit weight (g), fruit length (cm), fruit diameter (cm), fruit stalk length (cm), total soluble solids (TSS) (%), number of seeds per fruit, seed weight (g), seed length (cm), fruit moisture (%) and fruit dry matter (%), were recorded for 30 fruits and leaves with three replications for two seasons 2013–2014. These traits were measured in individual plants and were averaged for each of the 30 genotypes. The quantitative traits, related to the length and width of the leaf, fruit and seed, were measured with an accuracy of 0.1 mm by using a vernier calliper. The fruit and seed weights were measured by using analytical balance (model JKH-500, Jadever Co.) with a sensitivity of ± 0.01 g. The TSS content was determined by using juice samples of fruit pulp with a handheld refractometer (pocket PAL-1 ATAGO Corporation, Tokyo, Japan) at room temperature. Data analysis was performed by using SPSS software (version 6.1.).[Citation12] Mean values recorded for each parameter were used for statistical analyses with SPSS software. For each factor, a principal component with a value more than 0.50 was considered as being significant. Scatter plot was prepared according to the principal component (PC1 and PC2) by using palaeontological statistics (PAST) software.[Citation13] Polymorphism information content (PIC) (%) was determined as an indicator of variability, which is the ratio of the standard deviation to the mean (×100).
Table 1. Studied genotypes and their collection area in Iran.
DNA extraction and random amplified polymorphic DNA (RAPD) analysis
Genomic DNA was extracted from young leaves based on the method described by Doyle and Doyle, with slight modifications.[Citation14] Briefly, 100 mg of fresh leaf tissue was ground in liquid nitrogen. The ground tissue was suspended in 600 µL of isolation buffer (2%, w/v, hexadecyl trimethyl ammonium bromide (cetyl trimethylammonium bromide, CTAB), 1.4 mol/L NaCl, 20 mmol/L Ethylenediaminetetraacetic acid (EDTA, pH 8.0), 100 mmol/L Tris-HCl (pH 8.0), 1% (w/v) polyvinylpyrrolidone (PVP-40) and 1% (v/v) 2-mercaptoethanol and incubated for 60 min at 65 °C with occasional mixing by inversion. The mixture was allowed to cool down to a room temperature and a 1:1 chloroform:isoamyl alcohol (24:1) was added. The tubes were mixed well and incubated at room temperature for 20 min with occasional inversion. The tubes were then centrifuged (Labnet Prism-R C2500-R Refrigerated Microcentrifuge, USA) at room temperature for 15 min (13,000 rpm) and the supernatant was transferred to a separate tube. Subsequently, DNA was precipitated from the aqueous phase by adding 2/3 volume of cold isopropanol and centrifuged at 10,000 rpm for 10 min. The supernatant was then discarded. The pellet was washed with 0.2 mol/L sodium acetate and 70% (v/v) ethanol and dried in a vacuum oven (Fistreem International, Leicestershire, UK) at 30 ˚C for 15 min. The DNA pellet was dissolved in 100 μL distilled water.[Citation14] Twenty eight of Genset Oligos RAPD primers (SinaClon BioScience Co. Iran) (see below) were tested on three different genotypes and 10 of them produced clear and reproducible polymorphic bands. Polymerase chain reaction (PCR) amplification was carried out using the 10 RAPD primers. The PCR was carried out in a final volume of 15 µL containing 3 µL DNA template (10 ng/µL), 1.5 µL 10X PCR buffer (pH 8.0), 0.45 µL MgCl2 (50 mmol/L), 1 µL dNTPs (10 mmol/L), 2 µL primers (10 pmol/L), 0.3 µL Taq DNA Polymerase (5 U/µL) and 6.75 µL sterile distilled water. The PCR programme was as follows: one cycle for 5 min at 94 °C, for initial denaturation, followed by 40 cycles. Each cycle consisted of a denaturation step at 94 °C for 1 min, an annealing step at 37 °C for 1 min and an extension step at 72 °C for 2 min, followed by a final extension at 72 °C for 7 min. Before loading the PCR product, 5 µL of loading buffer (pH 8.0) was added to each sample. The PCR-amplified products were separated by electrophoresis (Biostep, HU20, UK) on 1.5% (w/v) agarose gels in Tris-borate-EDTA (TBE) buffer (pH 8.0) at 80 V for 120 min. The gel was stained in an ethidium bromide solution for 25 min, destained in distilled water and photographed under UV light with a Gel Doc system. A 100-bp DNA ladder (SinaClon BioScience Co. Iran) was used as a molecular weight size marker in the side lanes of each gel. Each band was scored as present (1) or absent (0) and data were analysed with the numerical taxonomy and multivariate analysis system (NTSYS-pc programme ver. 2.00) software package.[Citation15] PIC values were calculated according to Smith et al.[Citation16] The Jaccard genetic similarity coefficient values were used to visualize the genetic relationship among the genotypes and the distance matrix was used for cluster analysis. Dendrogram was constructed through sequential agglomerative hierarchic non-overlapping (SAHN) clustering program by the unweighted pair-group method with arithmetic averages (UPGMA), using NTSYS-pc software.
Results and discussion
Morphological characteristics
A wide variation was observed in most traits of the studied wild hawthorn genotypes. The mean, maximum and minimum values, as well as standard deviations and phenotypic diversity index of the traits, are presented in . High variability among genotypes was found for most of the traits, but fruit weight, fruit length, fruit diameter, area and length of leaf variables were the characteristics with the highest variations. Both leaves and stipules characteristics are frequently used in the taxonomy of the genus. There are three types of leaves.[Citation17,Citation18] The average fruit weight varied among different species of hawthorn genotypes. The lowest and highest fruit weights belonged to C. pentagyna (0.33 g) and C. pontica (3.57 g), respectively. Higher fruit weight along with higher flesh amount are the most important and desirable fruit characteristics in hawthorn breeding programmes.[Citation19] In previous studies, the fruit length and width of hawthorn genotypes in Turkey were found to range from 7.96 to 23.9 mm and from 8.00 to 22.78 mm, respectively.[Citation20–23] Taxonomic problems in the genus Crataegus and the description of a large number of new species and nothospecies during the last century by Old World botanists are the result of several factors. These factors are hybridisation,[Citation24] introgression, polyploidy and probably apomixis, which may occur and cause difficulties in identifying species.[Citation3,Citation4,Citation25] Grant [Citation26] explains the correlation between polyploidy and various factors, including climate, latitude, altitude, type of habitat, life form, breeding system, hybridity, cell size, chromosome size, chromosome structure, sex chromosome mechanism and genotype.[Citation26] Within this genus, inherently variable species, such as C. monogyna, occur, in which different types of leaf blades can be found. This is a feature, which is considered to be of taxonomic significance in other species.[Citation3]
Table 2. Minimum, maximum and mean values, as well as standard deviations and phenotypic diversity index of the traits in hawthorn genotypes.
Principal component analysis
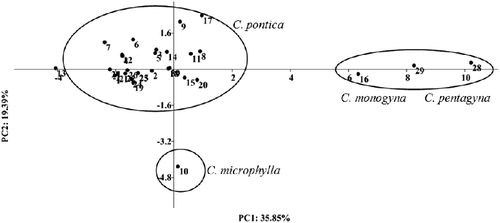
Principal component analysis (PCA) was used to identify the most significant traits in the data-set. The aim of PCA is to determine the main factors and effective parameters to discriminate among accessions. The correlations between the original traits and the first five principal components are shown in . The results from the PCA indicated that 85.05% of the observed variability was explained by the first five components. The first two components explained about 55.24% of the total achieved variability. PC1 and PC2 were represented mainly from fruit weight, fruit length, fruit diameter, fruit moisture, fruit dry matter, leaf length, leaf area, leaf width, petiole length, number of leaves per node, seed weight, seed length and fruit stalk length, which were predominant in the first two components and accounted for 55.24% of the total variance. According to Núñez-Colín et al.,[Citation27] leaf characteristics were the most useful in defining and comparing Crataegus germplasm sources.[Citation27] Scatter plot was prepared, based on the PC1and PC2 that reflected the relationship among hawthorn genotypes in terms of phenotypic resemblance and morphological traits ().
Table 3. Eigenvalues and cumulative variance for five major factors obtained from PCA and significant parameters within each component for hawthorn genotypes.
Molecular analysis
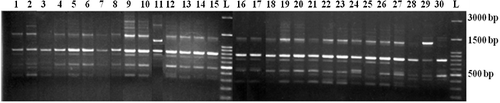
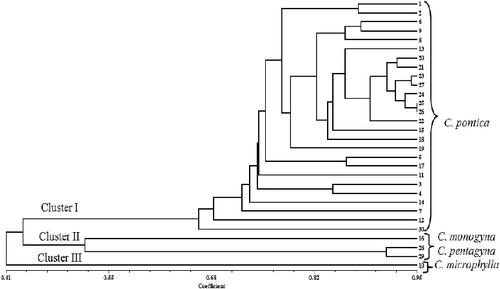
Ten RAPD primers reproducibly and strongly amplified 69 fragments, 58 of which were polymorphic. The size of the amplified fragments ranged from 180 to 2700 bp. The number of the observed polymorphic alleles for each locus ranged from 4 to 8, with an average of 5.8 alleles per locus (). The proportion of polymorphic fragments in the profile generated by each primer ranged from 66.67% (Oligo_16 and Oligo_339) to 100% (Oligo_32 and Oligo_35). The PIC was observed to be the highest (0.79) in the Oligo_32 locus, whereas the Oligo_339 locus had the lowest value of 0.64 among the RAPD primers. The allelic distribution of Oligo_35 is shown in . The Jaccard's genetic similarity coefficient ranged from 0.12 to 0.95 among the genotypes. The lowest genetic distance (0.12) was found to be between 8 and 29 accessions, which belonged to C. pontica and C. pentagyna species, whereas the highest genetic distance (0.95) was revealed to be between 25 and 26 accessions from C. pontica species. The cophenetic correlation coefficient showed high correlation (r = 0.83) between the RAPD similarity matrix and the cophenetic matrix, indicating a good representation of the molecular relationship among the genotypes. Dendrogram, constructed by using the UPGMA method, based on RAPD banding patterns, revealed high genetic variation among the genotypes. At a similarity coefficient of 0.46, UPGMA cluster analysis divided the genotypes into three major groups (). Overall, this cluster was similar to that of the PCA analysis-based morphological data. For example, there were three groups on the dendrogram and scatter plot, revealed by principal components analysis. The first main cluster included all of the 26 C. pontica genotypes (Cluster I). The second main cluster included the remaining three genotypes, belonging to C. monogyna and C. pentagyna (Cluster II). The third cluster included C. microphylla (Cluster III). The results from the cluster analysis supported the results from the scatter plot and the accessions were distributed in three clusters. In most cases, the high differentiation among populations was caused by factors, such as breeding system, isolation of populations, seed and pollen dispersal distance. The high differentiation might be a result from a habitat fragmentation, which has led to the isolation of populations, the decreasing of their size and the limitation of gene flow among them. Similar results were obtained in Italian C. monogyna.[Citation28] Dickinson et al. [Citation29] suggested that a better understanding of the modes of reproduction and the uses of flow cytometry and molecular markers would help us study the Crataegus taxonomy in greater detail.[Citation29] Our results indicated that RAPD markers can be successfully used to study the molecular relatedness of Crataegus species. There are many researches, reporting the successful applications of RAPD markers for the characterization of fruit accessions, such as olive [Citation30,Citation31] and hawthorn.[Citation28,Citation32–34] In all these studies, the molecular markers appear to be a reliable method for the analysis of genetic relationships within fruit trees.
Table 4. Data obtained from the RAPD primers used with the studied hawthorn accessions.
Conclusions
In the present study, we attempted to characterize some hawthorn accessions, which were collected from Iran, by using both morphological characteristics and molecular data. Due to polymorphism, hybridization and apomictic breeding strategies, the species of this genus show great variations in populations. In this study, the average fruit weight among different species of hawthorn genotypes varied. The difference in fruit weights and dimensions of germplasm under the same geographical conditions may be a result of genotypic effects. This investigation clearly indicated that a wide biodiversity occurred among hawthorn germplasms found in Iran. Fruit weight, fruit length, fruit diameter, area and length of leaves highly varied among genotypes. Moreover, since commercial hawthorn cultivars do not exist, these results could be important to give more information about the use of these genotypes as breeding material in future traditional breeding or advanced biotechnology studies.
Acknowledgments
The authors gratefully acknowledge the financial support for this work provided by Ilam University, Iran.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kumar D, Arya V, Bhat ZA, et al. The genus Crataegus: chemical and pharmacological perspectives. Br J Pharmacogn. 2012;22:1187–1200.
- Judd WS, Campbell CS, Kellogg EA, et al. Plant systematic – a phylogenetic approach. Sunderland (MA): Sinauer Associates; 1999.
- Christensen KI. Revision of Crataegus sect. Crataegus and nothosect. Crataeguineae (Rosaceae-Maloideae) in the Old World. Syst Bot Monograms. 1992;35: 1–199. doi:10.2307/25027810.
- Albarouki E, Peterson A. Molecular and morphological characterization of Crataegus L. species (Rosaceae) in southern Syria. Bot J Linn Soc. 2007;153:255–263.
- Byatt JI. Hybridisation between Crataegus monogyna; Jacq. and C. laevigata (Poiret) DC in south-eastern England. Watsonia. 1975;10:253–264.
- Depypere L, Mijnsbrugge KV, De Cock K, et al. Indigenous species of Crataegus (Rosaceae-Maloideae) in Flanders (Belgium). An explorative morphometric study. Belg J Bot. 2006;139(2):139–152.
- Dickinson TA, Campbell CS. Population structure and reproductive ecology in the Maloideae (Rosaceae). Syst Bot. 1991;16:350–362.
- Krawczyk U, Pteri G, Kery A. HPLC analysis of procyanidins in Crataegus extract. Arch Pharmazie. 1991;324:97–99.
- Dirr MA. Manual of woody landscape plants: their identification, ornamental, characteristics, culture: propagation and uses. Champaign (IL): Stipes Publishing; 1998.
- Archak S, Gaikwad AB, Gautam D, et al. Comparative assessment of DNA fingerprinting techniques (RAPD, ISSR and AFLP) for genetic analysis of cashew (Anacardium occidentale L.) accessions of India. Genome. 2003;46:362–369.
- Rahmani MS, Shabanian N, Khadivi-Khub A, et al. Population structure and genotypic variation of Crataegus pontica inferred by molecular markers. Gene. 2015;572:123–129.
- Norusis M. SPSS professional statistics 6.1. Chicago (IL): SPSS; 1994.
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9.
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15.
- Rohlf FJ. NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2. 1. Setauket (NY): Exeter Software; 2000.
- Smith, JSC, Chin ECL, Shu H, et al. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet. 1997;95:163–173.
- Dickinson TA. Topodeme differentiation in Ontario taxa of Crataegus (Rosaceae: Maloideae) leaf morphometric evidence. Can J Bot. 1986;64:2738–2747.
- Smith PG, Phipps JB. Studies in Crataegus (Rosaceae, Maloideae) IX. Short-leaf heteroblasty in Crataegus crus-galli sensu lato. Can J Bot. 1984;62:1775–1780.
- Ercisli S. A short review of the fruit germplasm resources of Turkey. Genetic Resour Crop Evol. 2004;51:419–435.
- Asma BM, Birhanlı O. Selection studies of wild growing hawthorns around Malatya. IV. Turkish National Horticultural Symposium; 2003 Sep 8–12; Antalya, Turkey.
- Balta MF, Celik F, Turkoglu N, et al. Some fruit traits of hawthorn (Crataegus spp.) genetic resources from Malatya, Turkey. Res J Agric Biol Sci. 2006;2:531–536.
- Turkoglu N, Kazankaya A, Sensoy RI. Pomological characteristics of hawthorn species found in Van Region. J Agric Sci. 2005;15:17–21.
- Yanar M, Ercisli S, Yilmaz KU, et al. Morphological and chemical diversity among hawthorn (Crataegus spp.) genotypes from Turkey. Scientific Res Essays. 2011;6(1):35–38.
- Phipps JB. A review of hybridization in north American hawthorns – another look at ‘the Crataegus problem’. Ann Missouri Botanical Garden. 2005;92:113–126.
- Lippert W. Crataegus. In: Hegi G, Conert HJ, Scholz H, editors., Illustrierte Flora von Mitteleuropa. Band 4. Angiospermae-dicotyledones 2.-(3). 2nd ed. Berlin: Blackwell Wissenschafts-Verlag; 1995. p. 426–445.
- Grant V. Plant speciation. New York (NY): Columbia University Press; 1981. Chapter 24, Factors promoting polyploidy; p. 307–323.
- Núñez-Colín, CA, Nieto-Ángel R, Barrientos-Priego AF, et al. Variability of three regional sources of germplasm of tejocote (Crataegus spp.) from central and southern Mexico. Genet Resour Crop Evol. 2008;55:1159–1165.
- Ferrazzini D, Monteleone I, Belletti P. Small-scale genetic diversity in one seed hawthorn (Crataegus monogyna Jaq.). Eur J For Res. 2008;127:407–414.
- Dickinson TA, Lo E, Talent N. Polyploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications. Plant Syst Evol. 2007;266:59–78.
- Ganino T, Beghe D, Alenti S, et al. RAPD and SSR markers for characterization and identification of ancient cultivars of Olea europaea L. in the Emilia region, Northern Italy. Genet Resour Crop Evol. 2007;54:1531–1540.
- Taamalli W, Geuna F, Banth R, et al. Agronomic and molecular analyses for the characterization of accessions in Tunisian olive germplasm collections. Electron J Biotechnol. 2006;9:467–481.
- Dai H, Guo X, Zhang Y, et al. Application of random amplified polymorphic DNA and inter-simple sequence repeat markers in the genus Crataegus. Ann Appl Biol. 2008;154:175–181.
- Serçe S, Şimşek O, Toplu C, et al. Relationships among Crataegus accessions sampled from Hatay, Turkey, as assessed by fruit characteristics and RAPD. Genet Resour Crop Evol. 2011;58:933–942.
- Yilmaz KU, Yanar M, Ercisli S, et al. Genetic relationships among some hawthorn (Crataegus spp.) species and genotypes. Biochem Gen. 2010;48:873–878.