ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterium that is resistant to a large group of beta-lactam antibiotics. Rhazya stricta is a local shrub that grows naturally as a normal flora and is used as a medicinal plant by several nations for a lot of infectious diseases, caused by microorganisms. Therefore, the effect of the plant against different genotypes of methicillin-resistant S. aureus was tested in the present study. Molecular identification was done for the medical sampling of 44 MRSA and biodiversity approaches were applied to detect the mecA gene. The 16S rRNA genes analysis was performed for the construction of a phylogenetic tree. Later on, the antimicrobial effect of the plant leaves’ water extract was tested on different genotypes. MecA gene appeared in all isolates, except in methicillin-susceptible Staphylococcus aureus. The selected MRSA 16S rRNA sequences were sent to GenBank and six accession numbers (KM893010, KM893011, KP091274, KP091275, KP137513 and KP137514) were acquired. Also, an evolutionary analysis of these strains was done and a phylogenetic tree was constructed. Plant extracts showed that the interaction between pathogens and drugs is more efficient in a liquid environment than in a solid one.
Introduction
The spread of microorganisms and their antibiotic resistance is a great threat to the worldwide medical community. Methicillin-resistant Staphylococcus aureus (MRSA) is a microorganism that causes concerns across the world. MRSA, by definition, is any strain of S. aureus that is resistant to a large group of antibiotics, called beta-lactams, which include penicillin and cephalosporin.[Citation1] MRSA emerged as a nosocomial pathogen in the early 1960s. At that time, most of the occurrences were sporadic outbreaks, but during the 1970s, an increasing number of large hospital outbreaks were reported in Europe and in many other countries, including USA, Japan and Australia. MRSA is the causative agent of many infectious diseases of the skin, soft tissues, respiratory tract, bones and joints, surgical wounds, urinary tract and bloodstream. These infections are difficult to treat because of the bacterial resistance to commonly used antibiotics.[Citation2] The increased prevalence of MRSA has been reported as a major cause of morbidity and mortality in hospitals for several decades.[Citation3] It is of utmost importance to know the prevalence of MRSA in any environment, because of the public health concerns and the threat posed by MRSA infections.[Citation4]
There are limited choices of antimicrobial agents for the treatment of many serious life-threatening infections, caused by MRSA, leading to prolonged stays of victimized individuals in the hospitals and increased care costs.[Citation5] Alternative therapeutic solutions, such as medicinal plants for the treatment of infections and diseases caused by resistant strains, may solve the problem of antibiotic drawbacks. Medicinal plants can also be used alongside antibiotics, in certain cases, as well as alone, depending on the underlying causes and types of diseases. This may consequently lead to the reduction and rationalization of the antibiotics usage. The use of medicinal plants have originated a long time ago for the treatment of infections and diseases.[Citation6] Many of the medicinal plants that grow naturally in the Kingdom of Saudi Arabia were used in the folkloric medicine to treat microbial infections and diseases.[Citation7,Citation8] Therefore, researches and studies of these local plants are essential to reveal more information regarding their therapeutic benefits and potential side effects. Information about the side effects would prove to be vital in the treatments’ standardization based on these medicinal shrubs. Notably, producing drugs from such plants will cost much lesser than producing and manufacturing a new antimicrobial agent.[Citation9]
Rhazya stricta is a local shrub that grows naturally as a normal flora in many regions of the world. This plant is known to have been used as a medicinal plant from a long time by several nations for the treatment of numerous diseases and microbial infections.[Citation10,Citation11] Later on, many studies and researches proved the positive antimicrobial effects of R. stricta against many species and strains of microorganisms.[Citation12–16] R. stricta is a plant, enriched with chemical compounds and secondary metabolites, but the therapeutic activities of most of these compounds are not yet discovered. The leaves of this plant are rich in alkaloids, glycosides, triterpenes, tannins and volatile bases.[Citation12–15,Citation17,Citation18]
The main objective of the present study was to identify the antimicrobial effect of R. stricta's plant extracts against the genotypes of resistant pathogens. The study depicts the molecular screening as a better method for the rapid identification and detection of isolates, as compared to the classical methods. The study covers Jeddah region in Saudi Arabia, due to the wide spreading of these pathogens in the Saudi communities and health care facilities.[Citation19,Citation20]
Materials and methods
Sampling
Forty-four isolates were collected by means of professional medical sampling by the staff of King Fahd Hospital, Jeddah, Saudi Arabia in the period between 1st of May and 30th of July, 2013. These isolates were characterized using biochemical tests, such as Gram staining, tests for oxidation/fermentation, production of acid from carbohydrates, hydrolysis of gelatin, Voges–Proskauer, indole production, methyl red and citrate production. This was performed according to Bergey's Manual of Systematic Bacteriology taxonomy.[Citation21] All media and chemicals used during the biochemical identification were from Thermo Scientific. Then the isolates were subcultured on blood agar (Thermo scientific) in the hospital laboratory. For better preservation, the isolates were subcultured in tryptic soy broth (Thermo Scientific) and mixed in microtubes with glycerol in a ratio of 1:1. Then they were stored at –80 °C until further applications. The reference strains MRSA ATCC 43330 (positive control) and methicillin-susceptible Staphylococcus aureus (MSSA) ATCC 25923 (negative control) were provided by the Microbiology laboratory at the Department of Biology, Faculty of Sciences at King Abdulaziz University.
Plant materials and extraction
R. stricta leaves were collected from Hadda, the old Jeddah Makkah road in Saudi Arabia (N 21.441012 - E 39.530714). The plant materials were collected in sterile plastic bags, stored in an ice box and delivered directly to the Department of Biology, Faculty of Sciences at King Abdulaziz University. The leaves were washed with sterile water for the removal of dust particles and 96 g were weighted to prepare a stock solution. For the preparation of the stock solution, the leaves were blended with 1 L of sterile distilled water and left overnight in a shaker. The next day, the extract was filtered through a sterile Whatman filter paper grade 1 and then was filtered again through a 0.22-μm sterile membrane filter (Sigma). The extract was kept in the refrigerator for one week for further work.[Citation22]
Molecular approaches
DNA was extracted from all 44 isolates, together with the positive and negative control reference strains, as described by Kalia et al.[Citation23] A polymerase chain reaction (PCR) amplification of the extracted DNAs was performed to identify the mecA genes and to confirm the classical microbiological diagnosis of the bacterial isolates, as described by Suleiman et al.[Citation24] The 16S rRNA genes analysis was applied for identifying the molecular diversity and constructing the isolates’ phylogenetic tree, as described by Shair.[Citation25] The mecA gene PCR was carried out in a 25-μL reaction mixture containing about 100 ng genomic DNA, 0.5 μmol/L of each primer (forward primer: 5'-AAAATCGATGGTAAAGGTTGGC-3' and reverse primer: 5'- AGTTCTGCAGTACCGGATTTGC-3'), 1.00 U of Taq DNA Polymerase, 2.5 μL of 10 × PCR assay buffer (1.5 mmol/L MgCl2), 100 μmol/L of each dNTPs. The amplification was performed with a pre-programmed MultiGene Thermal Cycler (Labnet International Inc., New Jersey, USA) and was set for 35 cycles after an initial denaturation cycle for 5 min at 95 °C. Each cycle consisted of denaturation step at 95 °C for 30 s, annealing step at 65 °C for 1 min and extension step at 72 °C for 1 min, followed by an extension step for 5 min at 72 °C, which was performed in the final cycle. Also, PCR for 16S rRNA gene was carried out in a 25-μL reaction mixture containing about 100 ng genomic DNA, 0.5 μmol/L of each primer (forward primer: 16F27:5'-AGAGTTTGATCCTGGCTCAG-3' and reverse primer: 16R1525 5'-AAGGAGGTGATCCAGCCGCA-3'), 1.00 U Taq DNA Polymerase, 2.5 μL 10 × PCR assay buffer (1.5 mmol/L MgCl2), 100 μmol/L of each dNTPs. The amplification was performed with a pre-programmed MultiGene Thermal Cycler (Labnet International Inc., New Jersey, USA) and was set for 30 cycles after an initial denaturation cycle for 5 min at 95 °C. Each cycle consisted of denaturation step at 95 °C for 30 s, annealing step at 65 °C for 30 s and extension step at 72 °C for 1 min, followed by an extension step for 10 min at 70 °C in the final cycle. The products from the 16S rRNA gene and mecA gene PCR were analyzed by electrophoresis (Sigma-Aldrich) in 1% agarose gel stained with ethidium bromide. DNA ladder (1 kb, Cleaver scientific) was used for determining the sizes of the bands. The gels were photographed by the Lab image analyzer software version 2.7.0. The 16S rRNA genes’ PCR products were then sent to Macrogen, Seoul in Korea for sequencing.
PCR product sequencing and basic local alignment search tool (BLAST) analysis
The results from the sequencing data were compared with all microbes’ sequences in GenBank by using the BLAST programme (http://www.ncbi.nlm.nih.gov/BLAST/). The sequences were sent to GenBank for attaining an accession number. Based on the 16S rRNA genes, the molecular evolutionary analyses of these strains were conducted by using 10 different S. aureus strains from GenBank (http://www.ncbi.nlm.nih.gov). The evolutionary distances were computed using the maximum composite likelihood method.[Citation26] The used units were the number of base substitutions per site. The evolutionary analyses and phylogenetic tree were constructed using MEGA version 4.[Citation27]
Antimicrobial tests
The antimicrobial activity test against S. aureus isolates was performed by using the disc diffusion method, according to the Clinical and Laboratory Standards Institute guidelines.[Citation28] The bacteria, used in this study, were MRSA S. aureus (ATCC 43330), MSSA S. aureus (ATCC 25923) and six clinical isolates that were obtained from the Laboratory of Microbiology, Faculty of Sciences at King Abdulaziz University. The dried plant extracts were dissolved in sterile distilled water to a final concentration of 6 (commonly used dose), 12, 24 and 48 g/L, and were sterilized by filtration through a 0.2-µm membrane filter (Whatman, USA).
The bacterial inoculum was uniformly spread by using a sterile cotton swab on a sterile Petri dish, containing Mueller–Hinton agar (Sigma-Aldrich). About 20 µL of the plant extract with different concentrations (6, 12, 24 and 48 g/L) were pipetted onto a sterile paper disc and transferred to a plate containing a bacterial culture. Then these plates were incubated for 24 h at 36 ± 1 °C, under aerobic conditions. After incubation, a confluent bacterial growth was observed. The inhibition of the bacterial growth was measured in mm. For the Mueller--Hinton broth (Sigma-Aldrich) experiments, freshly prepared cultures containing 6 g/L concentration of plant extracts were inoculated with bacterial isolates and incubated for one night at 37 °C.
Results and discussion
Molecular diversity and phylogeny
Around, 1500 bp of the 16S rRNA gene were amplified by using the specific primers. Selected MRSA 16S rRNA sequences were sent to the GenBank and six accession numbers (KM893010, KM893011, KP091274, KP091275, KP137513 and KP137514) were acquired. A phylogenetic tree of six selected MRSA sequences (KM893010, KM893011, KP091274, KP091275, KP137513 and KP137514) was constructed against 10 different S. aureus strains from GenBank (http://www.ncbi.nlm.nih.gov) by using the MEGA version 4, as shown in . The data showed that the six selected S. aureus (MRSA) 16S rRNA sequences have molecular relationships with many of the S. aureus strains from GenBank.
Figure 1. Comparative phylogenetic analysis of S. aureus (KM893010, KM893011, KP091274, KP091275, KP137513, KP137514) with other S. aureus strains from GenBank.
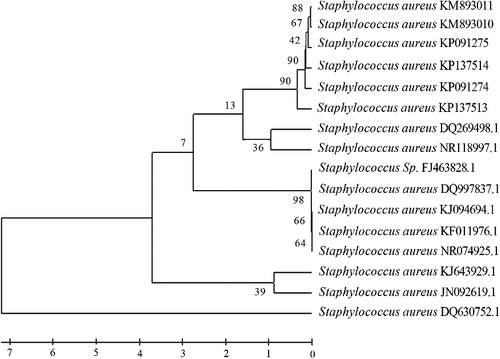
Many of the S. aureus strains were in the same cluster with the six selected MRSA and the 16S rRNA sequences. In this context, the results showed that the six S. aureus strains identified in the same group shared a high genetic similarity with each other, whereas they shared different genetic similarities with the other S. aureus strains from GenBank. The group containing S. aureus DQ269498.1 and NR_118997.1 shared the highest genetic similarity with the six S. aureus strains identified in the present study. S. aureus is an important pathogen, which causes several diseases in people and animals.[Citation29] These pathogenic effects of different S. aureus strains depend on several factors, such as colonization and adaptation patterns to different types of tissues and species in various microenvironments. Several methods have been utilized for the discrimination of different strains of S. aureus isolates. Molecular typing methods, such as detection of ribosomal RNA homology, have been proven to be of great value in discriminating these homogeneous strains. The molecular methods have also proved to be helpful in epidemiological studies, especially in measuring of bacterial isolates’ clonal relationships and in tracing the point of origin and the course of spread of the organism in the population.[Citation30]
Pathogen identification by using mecA gene
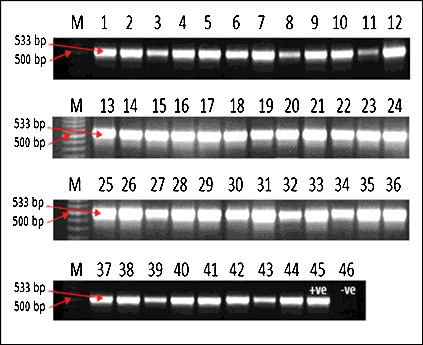
All 44 isolates, plus the positive control MRSA showed a mecA gene band with a molecular weight of 533 bp. The negative control MSSA did not show this band ().
The conclusions from the obtained results in the present study were similar to the ones recorded by Chan et al.[Citation31] The author evaluated the antibacterial activities against MRSA.[Citation31] In the study galloylated flavonol rhamnosides and quercetin, together with five known galloylated and non-galloylated flavonol rhamnosides, isolated from leaves of Calliandra tergemina (L.) Benth, were used as antibacterial agents against MRSA.[Citation31] By using the PCR method, all of the 44 tested isolates in the present study were found to be positive for mecA gene and also resistant to all classical microbiology diagnostic methods. This showed a good relationship between the molecular and classical approaches for the detection and diagnosis of such organisms and may also encourage the replacement of the classical methods with the faster and more accurate molecular approaches.
Antimicrobial effect of the plant extract on the bacterial isolates
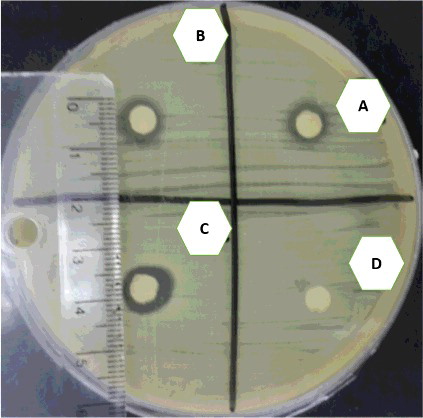
The results from the disc diffusion test on Mueller–Hinton agar did not go as expected for the commonly used dose (6 g/L) of R. stricta and there were no inhibition zones for the study isolates and control strains. The bacterial culture was also grown in the presence of plant extract in order to check the effect of the plant extract on the bacterial genome. The lack of growth in the liquid culture compared to the solid culture, with the same dose of the plant extract, will be discussed in the discussion section. To start thinking about how to use this medicinal plant and improve it later as a drug against MRSA and other multidrug-resistant organisms (MDRs), we tested higher doses of the plant extract (12, 24 and 48 g/L) against the bacterial isolates on Mueller–Hinton agar by using the disc diffusion assay. The results showed inhibition zones in the range between 8 and 10 mm, as shown in . The commonly used dose (6 g/L) did not show any inhibition zone for all 46 genotypes on the Mueller--Hinton agar. Also, no growth occurred in the Mueller–Hinton broth. On the other hand, the higher doses of extracts (12, 24 and 48 g/L) gave an inhibition zone that reached 10 mm on Mueller--Hinton agar.
The obtained results of the plant extract's antimicrobial effects were no different from the results obtained with other extraction methods used in other studies related to the same plant.[Citation14–16] Amin and Khan [Citation18] found that an organic fractionation of the plant has a positive effect against microorganisms when compared to the water extracts. The same results were obtained when this method was used in a study conducted for Gram-negative resistant bacteria.[Citation18] Conversely, a promising result obtained by Abadi et al. [Citation16] encourages the use of the plant water extract against study groups, because it had a strong effect on Neisseria meningitidis. Similar results were also reported in several researches [Citation13,Citation14,Citation17] by using water extracts of the plant against fungi. On the contrary, higher concentrations of the plant water extracts had a strong effect on our study group when applied in solid media, as well as when applied in a commonly used dose in liquid media. This could be explained with the fact that the efficiency of the drug would be higher in liquid environments than in solid ones, because of the direct interaction between drug and pathogen and also because of the ease of infiltration of the extract through the bacterial cell walls and membranes in the liquid medium. From here, we can state that the study regarding the organic fractions of extracts for this plant against MRSA and other MDRs is crucial and must be performed with chemo-informatics analysis to identify each component of the extract and to test each of them separately against resistant and newly emerging pathogens.
Molecular approaches are significant tools used for diagnostic purposes, which can be performed either alone, or along with classical diagnostic approaches. By using molecular markers and other molecular methods in diagnosing, all types of infections and diseases give quick and fine results and are also useful for proposing suitable drugs and therapies. Molecular and genomic tools are the best modern methods for genotyping and finding biodiversity, and must be applied to all local pathogens and species to fill gaps in the local and global gene banks. The use of medicinal plants must be scientifically researched to meet the pharmacological standards. Additional studies must be conducted for R. stricta and other medicinal plants to avoid all side effects, due to their crude utilization without any reliable studies. Further researches will help to enable the use of such plants in their legitimate pharmacological forms.
The results of plant extracts’ high concentrations in solid culture media extended the need to make a trial with more fractions of the desired dose. They also encourage the isolation of active compounds to analyze their effects on different pathogens. This will help to present suitable doses for communities in contrast to the legitimate pharmacological standards.
Conclusions
MRSA has become a worldwide public health problem, including in Saudi Arabia. As the infection-causing bacteria are resistant to first-line antibiotics, the treatment option remains a second or third choice of antibiotics, which are generally much more expensive. The present study investigated the antibacterial activity of R. stricta against clinical isolates of MRSA and revealed that the plant can be used as an alternative treatment therapy of infectious diseases caused by multidrug-resistant bacteria.
Acknowledgements
The team would like to express their thankfulness to the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia for the technical support and funding of this study, and would also like to thank the laboratory of King Fahd Hospital, Jeddah, Saudi Arabia for providing the samples of the study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Acton QA, editor. Community-acquired infection: new insights for the Healthcare professional. Chapter 5, Surgery; Atlanta (GA): Scholarly Editions; 2013.
- Gould IM. Cost of hospital acquired methicillin-resistant Staphylococcus aureus (MRSA) and its control. Int J Anti Microb Agents. 2006;28(5):379–384.
- Wilson P, Andrews JA, Charlesworth R, et al. Linezolid resistance in clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 2003;51(1):186–188.
- Robicsek A, Hacek DM, Fisher A, et al. Progression to MRSA infection in symptomatic carriers (AC) detected through a hospital-based universal surveillance and decolonization program. In: Program and abstracts of the forty-fourth annual meeting of the Infectious Diseases Society of America. Toronto: 2006. p. 12–15.
- Drew RH, Perfect JR, Srinath L, et al. Treatment of methicillin-resistant Staphylococcus aureus infections with quinupristin– dalfopristin in patient's intolerant of or failing prior therapy. J Antimicrob Chemother. 2000;46(5):775–784.
- Sharpe JN, Shively EH, Polk HC Jr. Clinical and economic outcomes of oral linezolid versus intravenous vancomycin in the treatment of MRSA-complicated, lower-extremity skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. Am J Surg. 2005;189(4):425–428.
- Paule SM, Hacek DM, Kufner B, et al. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus test before and during high-volume clinical use. J Clin Microbiol. 2007;45(9):2993–2998.
- Batanouny KH, Baeshin NA. Studies on the flora of Arabia: I. the Jeddah-Mecca Road, Saudia Arabia. Taeckholmia. 1978;9:67–81.
- Siegel JD, Rhinehart E, Jackson M, et al. The healthcare infection control practices advisory committee. Management of multidrug-resistant organisms in healthcare settings. Atlanta (GA): Centers for Disease Control and Prevention; 2006.
- Klevens RM, Edwards JR, Tenover FC, et al. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992–2003. Clin Infect Dis. 2006;42 (3):389–391.
- Ahmad S, Fatima K, Atiq-ur-Rahman. Antibacterial activity of Pakistani Rhazya stricta. Pak J Sci Ind Res. 2004;47(1):29–33.
- Kabli SA, Hadhoud AA, Baeshen MN. An epidemiological survey on ESBL producing bacteria genotypes and evaluating the antimicrobial effect of Rhazya stricta leaves extract upon them. Microbiol Res. 2012;3(2):66–73.
- Baeshin NA, Sabir JM. Some genetic studies on Aspergillus terreus Thom. Res Sci KAU. 1987;222:215–224.
- Baeshin NA, El-Twaty NH, Al-Hebshi AM. Evaluating the genotoxicity of Rhazya stricta leaves extract by the Saccharomyces cerevisiae auxotrophic mutants test. Egypt J Nat Tox. 2005;2(5):88–100.
- Gilani SA, Kikuchi A, Shinwari ZK, et al. Phytochemical, pharmacological and ethnobotanical studies of Rhazya stricta Decne. Phytother Res. 2006;21(4):301–307.
- Baeshin NA, Qari SH, Sabir JS, et al. Biochemical and molecular evaluation of genetic effects of Rhazya stricta (Decne) leaves extract on Aspergillus terreus. Saudi J Biol Sci. 2008;15(1):25–33.
- Abadi FJR, Abdulaziz AM, Hadhoud AA, et al. An epidemiological survey and evaluation of the antimicrobial growth effect of Rhazya stricta (Decne) leaves extract on different genotypes of Neisseria meningitidis. Egypt J Med Microbiol. 2007;20(2):77–68.
- Amin A, Khan MA. In vitro bactericidal and bacteriostatic potential of ingredients of traditional medicine obtained from Kacha Area (River Indus) district D.I.Khan, Kpk, against human bacterial pathogens. Pak J Bot. 2011;43(5):2613–2617.
- Baeshen NA, Elkady A, Abuzinadah OA, et al. Potential anticancer activity of the medicinal herb, Rhazya stricta, against human breast cancer. Afr J Biotechnol. 2012;11(37):8960–8972.
- Iyer AP, Baghallab I, Albaik M, et al. Nosocomial infections in Saudi Arabia caused by Methicillin resistance Staphylococcus aureus (MRSA). Clin Microbiol. 2014;3(3):2–5.
- Holt, SG, Kriey NR, Sneath PHA, et al. Bergey's manual of determinative for bacteriology. New York (NY): Williams and Wilkins; 1998.
- Iyer A, Kumosani T, Azhar E, et al. High incidence rate of methicillin-resistant Staphylococcus aureus (MRSA) among healthcare workers in Saudi Arabia. J Infect Dev Ctries. 2014;8(3):372–378.
- Kalia A, Rattan A, Chopra P. A method for extraction of high-quality and high-quantity genomic DNA generally applicable to pathogenic bacteria. Anal Biochem. 1999;275(1):1–5.
- Suleiman AB, Umoh VJ, Kwaga JKP, et al. Prevalence and antibiotic resistance profiles of Methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine mastitic milk in Plateau State, Nigeria. Int Res J Microbiol. 2012;2(8):264–270.
- Shair OMH. Actinomyces pyogenes isolates from Sheep: biochemical identification and confirmation by molecular method. Afr J Microbiol Res. 2012;6(6):1118–1124.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Natl Acad Sci. 2004;101:11030–11035.
- Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599.
- Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically (Approved standard-seventh edition). Wayne (PA): CLSI; 2006.
- Dubey A, Ghorui SK, Kashyap SK. Differentiation of Staphylococcus aureus strains based on 16s–23s ribosomal RNA intergenic space polymorphism. Indian J Biotechnol. 2009;8:276–279.
- Selander RK, Korhonen TK, Vaisanen-Rhen V, et al. Genetic relationship and clonal structure of strains of Escherichia coli in neonatal septicemia and meningitis. Infect immune. 1986;52:213–222.
- Chan EWL, Alexander IG, John OI, et al. Galloylated flavonol rhamnosides from the leaves of Calliandra tergemina with antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA). Phytochemistry. 2014;107:148–154.