?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Almost all optimization techniques are restricted by the problems' dimensions and large search spaces. This research focuses on a special hybrid method combining two meta-heuristic techniques, genetic algorithms (GA) and particle swarm optimization (PSO) that aims at overcoming this issue. This method investigates the potential impact of constraints (or the feasible regions) on the search pattern of GA and PSO. The proposed algorithm was applied for parameter estimation of batch fermentation process for alkaline protease production by Bacillus licheniformis in submerged culture. Furthermore, a comparison of proposed hybrid GA/PSO with pure GA and pure PSO was carried out. The results revealed that combination of these two meta-heuristic algorithms speeds up the search (about two-fold faster) in comparison to pure algorithms, since it benefits from synergy. Hence, the proposed method can be considered as an applicable method for parameter estimation of biological models in particular for large search space problems. Also, it was concluded that PSO has a slightly better performance and possesses better convergence and computational time than GA.
Introduction
Accurate estimation of the kinetic parameters of enzymes is an important part in the analysis of biochemical problems.[Citation1] Meta-heuristics are generic methods which offer good solutions, even global optima, within reasonable computing time. Thus, the use of meta-heuristics has received more and more attention.[Citation2] Different meta-heuristic methods have been applied to surmount the parameter estimation difficulties. Genetic algorithms (GAs), originally developed by Holland in 1975,[Citation3] are quite promising as a stochastic global optimization method.[Citation4] GA belongs to the larger class of evolutionary algorithms, which use an approach inspired by natural evolution, e.g. mutation, selection and crossover, to solve optimization problems.[Citation5] GA is identical to biological evolution in the principles of natural selection and the survival of the fittest.[Citation6] It is perhaps the most popular method used in parameter estimation problems.[Citation7] Particle swarm optimization (PSO), proposed by Kennedy and Eberhart in 1995, [Citation8] is also a meta-heuristic approach inspired by social models and swarming theory to find good solutions to optimization problems.[Citation9]
These optimization approaches can find global optima more quickly through cooperation and competition among the population of potential solutions of the search space even for complex optimization problems such as fermentation processes.[Citation10]
In recent years, both the GA and the PSO, as well as hybrid algorithms combining the two, have been applied to parameter estimation problems in the context of kinetic models of microorganism growth.[Citation7,Citation10–17]
GA has an advantage in exploration search and PSO has an advantage in sharing movement information between particles.[Citation18] Therefore in order to keep the advantages of both algorithms, combining between GA and PSO seems to be a logical method.
Also, many optimization problems are constrained with different conditions. Due to the existence of constraints, the optimization problems are more difficult than the unconstrained ones. Therefore, research on how to make the objective function find the optimal solution in the feasible region has great significance.[Citation19]
Penalty methods were developed in order to eliminate some or all of the constraints and add to the objective function a penalty term which prescribes a high cost to infeasible points.[Citation20] Therefore, in this work, growth kinetic parameter estimation has been carried out by a hybrid GA/PSO algorithm which investigates the potential impact of constraints (or the feasible regions) on the search pattern of GA and PSO. Finally, the result of the proposed hybrid algorithm has been compared to pure algorithms.
Materials and methods
Micro-organisms
The bacterial strain used in this study was isolated from soils collected at different locations in north of Iran. It was identified as Bacillus licheniformis (strain MG5), as described previously.[Citation21] The strain was maintained on nutrient agar slant and used throughout the study.
Fermentation conditions
The pre-culture medium was nutrient broth containing 2 g/L yeast extract, 5 g/L peptone, 5 g/L NaCl and 1 g/L beef extract, sterilized at 121 °C for 15 min.[Citation22]
Production of protease was carried out in 500-mL shake-flasks with 100 mL medium containing the following constituents: 20 g/L glucose, 7 g/L K2HPO4, 2 g/L KH2PO4, 0.2 g/L MgSO4.7H2O, 0.5 g/L NaCl, 1 g/L (NH4)2SO4 and 0.07 g/L CaCl2. Separately sterilized glucose was added into the medium as a carbon source just before inoculation. The above-mentioned pre-cultures were inoculated and the submerged cultures were incubated at 32 °C and 150 r/min in a shaker incubator.[Citation11]
Analytical procedures
Periodically, samples were withdrawn and the biomass concentration, residual glucose concentration and protease enzyme activity were determined. Bacterial growth was monitored by using a spectrophotometer at an optical density of 600 nm (OD600), which was then converted to cell dry weight (CDW) using a calibration curve. A standard graph was plotted for CDW versus absorbance for further estimation of CDW.[Citation22] Dinitrosallicylic acid was used for residual sugar assessment.[Citation23] Protease activity was determined as released tyrosine from the supernatants according to a modified Lowry method.[Citation24]
All experiments were conducted in triplicate and the experimental data given in this paper are arithmetic mean values of the triplicate experiments.
Problem formulation
Batch fermentation refers to a partially closed system in which most of the required components for micro-organism growth and maintenance are loaded before the process starts.[Citation15] The mathematical model of the batch fermentation process is presented by the following nonlinear differential equation system [Citation25]:
(1)
(1)
(2)
(2)
(3)
(3)
In the above equations, X, S and P are the biomass concentration, substrate concentration and product concentration, respectively (g/L). Xmax is the maximum biomass concentration (g/L). X0, S0 and P0 are the initial concentration of biomass, substrate and product, respectively (g/L), t is the current fermentation time (h), μmax is the maximum specific growth rate (h−1), YP/X is the product yield in terms of biomass (gP/gX) and k is the secondary coefficient of product formation or destruction (gP/g.h), YX/S is the biomass yield in terms of substrate (gX/gS) and ms is the maintenance coefficient (gS/gX.h).
EquationEquation (1)(1)
(1) is logistic equation that describes the dynamics of cell mass concentration whose growth is dependent on the availability of substrate and also on the total cell population. EquationEquation (2)
(2)
(2) is the Luedeking–Piret equation that describes the product formation and EquationEquation (3)
(3)
(3) is the modified Luedeking–Piret equation that describes the dynamics of substrate concentration.
Parameter estimation
In this study, the kinetic parameters estimated were { } as defined previously in the differential EquationEquations (1)
(1)
(1) –(Equation3
(3)
(3) ). The following bounds were used for the five kinetic parameters (n = 5):
. Each bound is divided to m sections; m is the same for the all kinetic parameters (at first m was set to 2)
(4)
(4)
(5)
(5) where
is the dth parameter,
and
are the lower and the upper bounds of the dth parameter, respectively. Also
denote the dth parameter bounded to the ith sub-bound of the parameter bound.
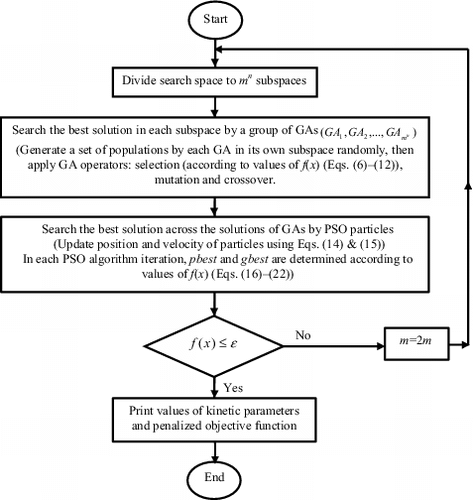
In the proposed hybrid GA/PSO algorithm, the search space is divided to mn subspaces (EquationEquation (5)(5)
(5) ). Each subspace consists of a vector of solutions named x {
, where each one of i1, i2, … and id are an integer in the range [Citation1,Citation10]}. Then a group of GAs
searches the best solution in each subspace. This means that a set of populations is generated by each GA in its own subspace randomly; each population introduces a set of potential solutions to the problem. The potential solution (individual) is coded as a binary vector, called a “chromosome”, the elements of which are called ‘genes’. In the GA algorithm, the population has chromosomes that represent candidate solutions; each chromosome is an n-dimensional real value vector where n is the number of optimized parameters. Therefore, each optimized parameter represents a dimension of the problem space.
One chromosome consists of five genes, coding for the parameters { }, respectively. One gene is coded by a binary string of 20 bits. Therefore, every possible solution of the parameters is simply represented by a binary string of 100 bits. Then, via decoding, the binary number of each gene in the chromosome is transformed to a decimal system and for obtaining simulated values of biomass, substrate and product, a set of nonlinear differential equations (EquationEquation (1)
(1)
(1) –(Equation3
(3)
(3) )) is solved by the fourth-order Runge–Kutta method (equation kinetic parameters are replaced with the ones estimated by GAs). Next, the value of the penalized objective function f(x) to be minimized is calculated:
(6)
(6)
Subject to
(7)
(7)
(8)
(8)
The penalized objective function f(x) is defined as
(9)
(9)
(10)
(10)
(11)
(11)
If and
,
and
will be zero. Therefore, the constraint does not affect f(x), but if the constraint is violated, that means
or
, f(x) will increase. where
(12)
(12) where p(x) is the penalty term; z is the number of experimental data items; Xexp, Sexp and Pexp are the experimental data for biomass (B. licheniformis), substrate (glucose) and product (alkaline protease); Xpred, Spred and Ppred are the predicted values for biomass, substrate and production with a given set of parameters;
is a vector of feasible solutions; F is the feasible region and W is the whole search space.
In each feasible region, after sorting the populations according to descending f(x) values, encoding is done and a genetic selection procedure is applied. Then two GA operators, mutation and crossover, are applied. When the final criterion (maximum number of generations: 100) is met, the GA operators are stopped.
Next, in the PSO algorithm, the population has mn particles. PSO particles search for the best solution across the solutions of GAs (from the previous step) in the whole search space. The position and velocity of particles are updated by applying an operator until particles can be expected to change towards the better solution. The updating procedure is defined by the following equations:
(13)
(13)
New velocity
(14)
(14)
New position
(15)
(15)
It should be noted that after decoding, the binary number of particles' position and velocity is transformed to a decimal system and for obtaining simulated values of biomass, substrate and product, a set of nonlinear differential equations (EquationEquation (1)(1)
(1) –(Equation3
(3)
(3) )) is solved by the fourth-order Runge–Kutta method (the equation kinetic parameters are replaced with the ones estimated by particles, in each iteration). Next, in each PSO algorithm iteration, the personal best and global best are determined according to the values of a penalized objective function similar to the previous step except that the search is done in the whole search space:
(16)
(16)
Subject to
(17)
(17)
(18)
(18)
(19)
(19)
(20)
(20)
(21)
(21) where
(22)
(22)
When the final criterion (maximum number of iterations: 100) is met, the PSO algorithm is stopped. Next, the solution is investigated. If the value of the penalized objective function (EquationEquation (16)(16)
(16) ) is less than
(
), values of parameters would be printed; otherwise, the number of feasible region in first step will be double (m = 2m) and the procedure is repeated. The flowchart of the hybrid GA/PSO is shown in
All calculations of hybrid GA/PSO were implemented in Matlab R2012b (MathWorks Inc., Massachusetts, USA).
Results and discussion
shows the comparison between simulated data by hybrid GA/PSO and experimental data in batch mode. The simulated data are obtained by solving EquationEquations (1)(1)
(1) –(Equation3
(3)
(3) ) by the fourth-order Runge–Kutta method (the equation parameters are replaced with the ones estimated by the hybrid GA/PSO method).
Figure 2. Experimental concentrations of X (٭), S (○) and P (◊) vs. hybrid GA/PSO simulated data (solid lines). Note: X, biomass (B. licheniformis); S, substrate (glucose); P, product (protease). Initial concentration of substrate 40 g/L.
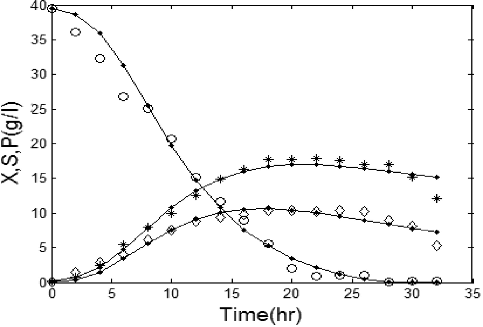
shows the parameter estimation results by the hybrid GA/PSO method. The performance of this method is evaluated by calculating the value of the penalized objective function (f(x)). The value of f(x) in the hybrid GA/PSO method was obtained . This proves the hybrid GA/PSO method to have a good ability to estimate kinetic parameters of alkaline protease production in batch mode.
Table 1. Kinetic parameters estimated by hybrid GA/PSO algorithm.
For comparison of the performance of the hybrid algorithm, we used pure GA and the pure PSO. shows the comparison between experimental data and simulated ones by GA and PSO, respectively.
Figure 3. Comparison of the experimental and simulated data of batch fermentation by using GA (A) and PSO (B). Experimental concentrations of X (٭), S (○) and P (◊) vs. simulated data (solid lines).
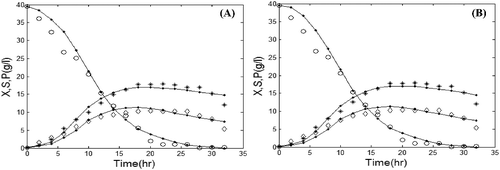
and show that all the three algorithms successfully model the dynamics of protease production in batch mode. Nevertheless, the values of the penalized objective function and the computation time that are presented in reveal that the hybrid GA/PSO fits the real experimental data more precisely than conventional algorithms.
Table 2. Comparison of performance across GA, PSO and hybrid GA/PSO algorithm.
We observed that our hybrid algorithm achieves results similar to the pure algorithms (the value of the objective function is close to these obtained by the pure GA and the pure PSO algorithms), but it uses less computation time (t = 48.34 s). The time used is about two times less compared to that used by the other ones.
Also the results showed that the computational effort required by PSO to arrive at such high quality solutions is less than the effort required to arrive at the same high quality solutions by the GA (t = 92.13 s for PSO and t = 104.92 s for GA). Furthermore, PSO has slightly better performance and possesses better convergence property than GA because PSO is easy to implement and there are few parameters to adjust. Thus, it must be concluded that the PSO approach is superior to the GA approach in this particular case.
Overall, the results indicate that both GA and PSO can be used in the parameter estimation of the biological models. In terms of computational effort, the PSO approach is faster, although it should be noted that neither algorithm takes what can be considered an unacceptably long time to determine their results. In our particular case, the hybrid GA/PSO technique gave successful results in parameter estimation because optimizers work jointly to efficiently locate quality design points better than what either of these could alone. The results prove that this hybrid algorithm can be considered a good and often superior technique for solving parameter estimation problem in biological models by fitting the simulation data with the corresponding experimental measurements.
Conclusions
Fermentation processes are very complex and thus it is often very difficult to draw a comprehensive picture of what actually takes place in a given fermentation process of interest. Therefore, the estimation of growth kinetic parameters in fermentation processes has become the primary objective for investigation of fermentation behaviour. In this study, a hybrid GA/PSO technique was successfully used for parameter estimation of simple kinetic models for protease production by using experimental data obtained from batch bacterial fermentation in submerged culture. The hybrid GA/PSO technique can be considered superior to pure GA or pure PSO because it optimizes the work jointly to efficiently locate quality design points better than either of these could alone. The proposed hybrid GA/PSO is also more effective in terms of execution time and solution quality, since it benefits from synergy. The results prove that this hybrid algorithm is a good and often superior technique for parameter estimation problem in biological models by fitting simulation data with the corresponding experimental measurements.
Acknowledgments
The authors would like to express their profound gratitude to Keivan Bolouri (Department of Industrial Engineering, Science and Research Branch, Islamic Azad University, Tehran, Iran) for his invaluable guidance and helpful comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Hemandez A, Ruiz MR. An EXCEL template for calculation of enzyme kinetic parameters by non-linear regression. Bioinform Appl Note. 1998;14:227–228.
- Roeva ON, Fidanova SS. Hybrid Bat algorithm for parameter identification of an E. coli cultivation process model. Biotechnol Biotechnol Equip. 2013;27:4323–4326.
- Holland J. Adaptation in natural and artificial systems. Ann Arbor (MI): The University of Michigan Press; 1975.
- Angelova M, Pencheva T. Tuning genetic algorithm parameters to improve convergence time. Int J Chem Eng. 2011; Article ID 646917. Available from: http://dx.doi.org/10.1155/2011/646917
- Karthikeyan P, Baskar S, Alphones A. Improved genetic algorithm using different genetic operator combinations (GOCs) for multicast routing in ad hoc networks. Soft Comput. 2013;17:1563–1572.
- Ahioglu S, Altinten A, Ertunc S, et al. Fuzzy control with genetic algorithm in a batch bioreactor. Appl Biochem Biotechnol. 2013;171:2201–2219.
- Sun J, Garibaldi JM, Hodgman C. Parameter estimation using metaheuristics in systems biology: a comprehensive review. IEEE/ACM Trans Comput Boil Bioinform. 2012;9:185–202.
- Kennedy J, Eberhart RC. Particle swarm optimization. In: IEEE International Conference on Neural Networks. Vol. 4; 1995 Nov 27–Dec 1; Perth. Piscataway (NJ): IEEE Service Center; 1995.
- Rada-Vilela J, Johnston M, Zhang M. Population statistics for particle swarm optimization: single-evaluation methods in noisy optimization problems. Soft Comput. 2015;19(9):2691–2716.
- Garlapati VK, Vundavilli PR, Banerjee R. Evaluation of lipase production by genetic algorithm and particle swarm optimization and their comparative study. Appl Biochem Biotechnol. 2010;162:1350–1361.
- Ghovvati M, Khayati G, Attar H, et al. Comparison across growth kinetic models of alkaline protease production in batch and fed-batch fermentation using hybrid genetic algorithm and particle swarm optimization. Biotechnol Biotechnol Equip. 2015;29(6):1216–1225.
- Rivera EC, Costa AC, Atala DI, et al. Evaluation of optimization techniques for parameter estimation: application to ethanol fermentation considering the effect of temperature. Process Biochem. 2006;41:1682–1687.
- Skolpap W, Nuchprayoon S, Scharer JM, et al. Fed-batch optimization of α-amylase and protease-producing Bacillus subtilis using genetic algorithm and particle swarm optimization. Chem Eng Sci. 2008;63:4090–4099.
- Abdullah A, Deris S, Mohamad MS, et al. A new particle swarm evolutionary optimization for parameter estimation of biological models. Int J Comput Inform Syst Ind Manag Appl. 2013;5:571–580.
- Benjamin KK, Emmanuel AN, David A, et al. Genetic algorithms using for a batch fermentation process identification. Appl Sci. 2008;8:2272–2278.
- Dutta JR, Dutta PK, Banerjee R. Modeling and optimization of protease production by a newly isolated Pseudomonas sp. using a genetic algorithm. Process Biochem. 2005;40:879–884.
- Calcada D. Modeling of the physiology of D. hansenii using population-based search methods for parameter estimation [dissertation]. Lisbon: Technical University of Lisbon; 2009.
- Nootyaskool S. The hybrid implementation genetic algorithm with particle swarm optimization to solve the unconstrained optimization problems. In: 4th International Conference on Knowledge and Smart Technology (KST); 2012 Jul 7–8; Chonburi. IEEE; 2012. doi:10.1109/KST.2012.6287739
- Kang LV, Yuanyuan MA. Modified PSO algorithm used in the solution of problems with constraints and its application. Int J Multimedia Ubiquitous Eng. 2014;9:97–108.
- Bergsma WP, Rapcsak T. An exact penalty method for smooth equality constrained optimization with application to maximum likelihood estimation. Eindhoven: Eurandom; 2005.
- Anvari M, Khayati G. Production and characterization of alkaline protease from Bacillus licheniformis sp. isolated from Iranian northern soils with ram horn hydrolysate. Trends Appl Sci Res. 2011;6:1206–1213.
- Anvari M, Khayati G. In situ recovery of 2,3-butanediol from fermentation by liquid–liquid extraction. J Ind Microbiol Biotechnol. 2009;36:313–317.
- Miller GL. Use of dinitrosalycilic acid reagent for determination of reducing sugars. Anal Chem. 1959;31:426–430.
- Meyers SP, Ahearn DG. Extracellular proteolysis by Candida lipolytica. Mycologia. 1977;69:646–651.
- Pazouki M, Najafpour G, Hosseini MR. Kinetic models of cell growth, substrate utilization and bio-decolorization of distillery wastewater by Aspergillus fumigatus UB260. Afr J Biotechnol. 2008;7:1369–1376.