?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Transcriptional activity of four defence-related genes (NPR1, PAL, defensin and PR5) in seven sunflower lines – resistant (MS-2161A), tolerant (MS-2039A) and susceptible (MS-2098A, MS-2091A, MS-2077A, MS-2067A and MS-1589A), infected with three broomrape populations (Tulcea, Romania; Soroca and Anenii Noi, Republic of Moldova), was studied in advanced stages of infection (90 days after sowing).
Obtained results revealed that resistant genotypes are characterized through higher stability in transcriptional activity of the studied genes. Thus, resistance could be associated with ability rapidly to maintain and recover a normal level of metabolism under more intensive stress factors. Also, it was established that expression of PR5 and defensin genes was altered and revealed considerable deviations in this phase of adaptation, while NPR1 and PAL mostly showed values at the level of the control group, which allows assuming that these genes, in the moment of sample collection and analysis, had normalized transcriptional activity, probably being involved in early responses.
Introduction
During the evolution plants have developed a large set of defence mechanisms against pathogens and herbivore insects including a plethora of passive pre-existing (structural barriers or some constitutively synthetized antimicrobial compounds, etc.) and active inducible responses (hypersensitivity response, phytoalexin biosynthesis, lignification and the reinforcement of the cell wall, production of pathogenesis-related proteins, etc.) that take place in the pest-colonized organ.[Citation1] Although defence responses can occur in the non-colonized organs and tissues of plants, e.g. systemic acquired resistance (SAR) – an inducible defence mechanism that is activated in the distal non-infected organs of a plant in response to a local infection with a pathogen.[Citation2]
According to the theory of adaptation process,[Citation3] immediately after plant exposure to stress, a reaction could be registered, which intensity is dependent to some degree of plant potential stress-resistance. After long exposure to stress, plants enter in adaptation stage, when changes in metabolism are insignificant. Thus, response reaction of living organisms to biotic and abiotic stress factors in plants includes following stages: irritation, damage and adaptation. In adaptation stage level of recovery of metabolic stability significantly differ within resistant, tolerant and susceptible plants.
At the cellular level plants respond to biotic and abiotic stresses by launching gene expression programs that contribute to adjustment of the physiological and metabolic processes to the new conditions and protection of cells against damage.[Citation4] Studies of gene expression in plants under biotic stress conditions [Citation5,Citation6] have provided comprehensive insights into transcriptional responses to a wide range of pathogens in different plant species.
At the molecular level, it was demonstrated that most of gene expression responses are temporary, transient and when presence of the stress factor persists, gene expression returns, after some time, to new steady-state levels that are closer to these characteristics in the unstressed organism. Stress responses show considerable differences among diverse plant pathosystems.[Citation4]
Previous studies performed on sunflower–broomrape pathosystem (in vitro co-culture) highlighted expression patterns of 11 defence-related genes (including PR5, defensin (PR-12; PDF), phenylalanine ammonia-lyase (PAL) and other) in roots of resistant and susceptible genotypes at 1, 8, 13, 17 and 21 days post-inoculation (d.p.i.). In inoculated roots of the resistant sunflower, some genes (PAL, cinnamate-4-hydroxylase and chalcone synthase) involved in the phenylpropanoid metabolic pathway, were induced. Also, it was detected up-regulation of transcriptional activity for some SA- and JA-responsive genes (PR5 and defensin, respectively).[Citation7]
Moreover, NPR1, Ha-PR5 and Ha-PDF gene expression was evaluated in sunflower hypocotyls under infection with downy mildew. NPR1 gene early and local activation was registered and associated with hypersensitive response and SAR triggering. The transcript levels of Ha-PDF and Ha-PR5 genes were increased in resistant genotypes, but lack of or delayed activation of these genes was observed in the case of susceptibility.[Citation8]
Although the defence responses, involving PR and other defence-related proteins, have been studied in details in model plant species such as Arabidopsis [Citation9,Citation10,Citation11] and rice,[Citation12,Citation13] to complete our understanding of resistance mechanisms in other plants, it is important to study defence responses in non-model plants. Furthermore, defence mechanisms are different within plant species, thus, each particular pathosystem should be investigated in details.
Thus, this study aimed to identify genes differentially expressed in the sunflower host plant during attack of three broomrape populations, collected from different geographical regions.
Attention was focused on expression of four genes NPR1, PAL, defensin and PR5 crucial for defence responses in plants infected with broomrape, in order to understand some of the adaptation and defence mechanisms which provides sunflower resistance to Orobanche cumana Wallr. in advanced stages of infection.
Materials and methods
Biological material
Seven cytoplasmic male sterility (CMS) sunflower lines (MS-2161A, MS-2098A, MS-2039A, MS-2091A, MS-2077A, MS-2067A and MS-1589A) created and offered by AMG-Agroselect Company were used as host plant biological material. To produce infection, three broomrape populations, collected from different geographical regions: two from Republic of Moldova (Soroca and Anenii Noi) and other one from Romania (Tulcea), were applied.
Growth conditions
Sunflowers were grown in a greenhouse at 18–20 °C during the day and 12–14 °C at night under a 14 h photoperiod. Seeds were planted in individual pots (volume 35–40 L), which contained artificially infected with broomrape seeds soil. Broomrape seeds were gathered from 100 plants grown in the same field. Inoculum density used in experiments was 150 mg of broomrape seeds per 1 kg of soil. The control samples were sowed in the same conditions, but on the soil without infection.
Sample collection
For gene expression analysis, leaves of three different sunflower plants in R5 developmental stage (90 days after sowing) [Citation14] were collected and immediately frozen in liquid nitrogen. Samples were collected from plants with aerial shoots – infected, plants grown on infected soil, but without aerial shoots at surface – non-symptomatic and without infection – control group.
Evaluation of infection
Frequency of attack was estimated using formula: , where N – number of attacked plants, Nt – total number of plants. Analysis of attack frequency highlighted virulence groups of pathogen and resistance of investigated genotypes. Plants with attack frequency 0%–5% are considered resistant (R); with F = 5%–20% were accepted to be tolerant (T); 20%–50% – susceptible (S) and these with frequency higher than 50% highly susceptible (SS).[Citation15]
RNA extraction and cDNA synthesis
The total RNA was extracted using TRI Reagent (Ambion, Applied Biosystems) from three different plants and mixed in a single bulked sample. Purified RNA samples were treated with DNAse I, RNAse-free (Thermo Scientific). First strand cDNA for each bulked sample (0.6 μg of RNA) was synthesized using RevertAid RT Kit (Thermo Scientific) following the manufacturer's instructions.
Gene expression analysis via qRT-PCR
Primers for quantitative real-time RT-PCR were designed using web based primer designing tool Primer3Web v.3.0.0.[Citation16] ().
Table 1. Genes and primers used in study.
Table 2. Phytopathological parameters estimation on investigated genotypes, artificially infected with three broomrape populations.
The mRNA expression levels of selected probes were analysed by quantitative real-time PCR (qPCR) using Maxima SYBR Green/ROX PCR Master Mix (Thermo Scientific) according to the manufacturer's protocols on DT-96 (DNA Technology, Russia). Each sample was analysed in three replicates performed in three different runs. To normalize the target gene expression, the ΔCt value was calculated, which presents difference between the Ct of the target gene and the Ct of sunflower actin (constitutive control) for the respective template according to Livak and Schmittgen (2001) [Citation17]:
Fold changes in gene expression were calculated as follows: 2−ΔΔCt.
Amplification of gene-specific products was confirmed by melting curve analysis, followed by agarose gel electrophoresis. Statistical analysis was performed according to Dospekhov (1985).[Citation18] Standard t-test was used for estimation of significant differences. Fold changes greater than 1.5 and lower than −1.5 were considered biologically significant.
Results and discussion
Sunflower (Helianthus annuus L.) is one of the most important crops in Republic of Moldova, being at third place after maize and wheat according to cultivated area.[Citation19] Sunflower is highly susceptible to the broad range of pathogens – fungi, oomycetes, insects, nematodes, etc., but in eastern Europe the greatest threat presents broomrape (Orobanche cumana Wallr.).[Citation20] For reasons of exaggerated extension of sunflower cultivated areas in RM (around 300.000 ha while scientists recommend optimum of 170.000 hectares) and violated crop rotation, broomrape attack risk increased in recent years. Simultaneously, it was determined that broomrape spreading significantly increased in the northern region of RM, being identified infected surfaces even in Soroca and Donduseni. Thus, research on screening of broomrape resistance, development and obtaining of resistant hybrids is of great interest and importance.
During this study, seven sunflower lines, which according to field experiments for several years showed under natural conditions resistance (MS-2161A), tolerance (MS-2039A) and susceptibility (MS-2098A, MS-2091A, MS-2077A, MS-2067A and MS-1589A) to broomrape, were used to analyse modification of the transcriptional activity of a set of genes during infection produced by three different broomrape populations (). These results were partly confirmed in controlled experiments model. These results were partly confirmed in controlled conditions experiments (greenhouse, laboratory, etc.).
The presence of O. cumana seedlings close/attached to sunflower roots altered expression of studied genes in both resistant and susceptible genotypes. The most resistant genotype MS-2161A showed the highest stability in transcriptional activity of studied genes. Significant changes were detected under influence of broomrape population collected from Tulcea, Romania.
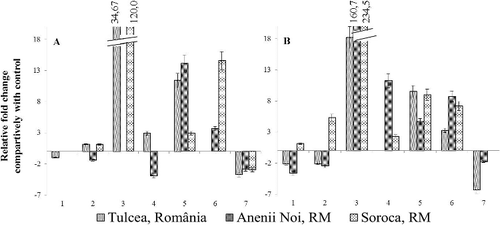
Another genotype, which manifested partial resistance – MS-2039A, similarly to MS-2161A was characterized by high stability of expression of studied genes with the exception of PR5 gene, which was strongly up-regulated (up to 35-fold in case of Tulcea population, up to 61-fold in case of Anenii Noi population and more than 120-fold in case of Soroca population) ().
Thus, resistance of plants is considered to be caused by the ability to maintain and recover a normal level of metabolism in a wider range of stress factor values intensity.[Citation3] Our findings regarding the most stable expression in resistant genotypes confirm this suggestion.
Comparatively, with resistant and tolerant genotypes, susceptible ones demonstrated higher deviations in transcriptional activity of investigated genes relatively to control group, which demonstrate instability of metabolic activity and worse adaptability to biotic stress.
In infected and non-symptomatic plants alterations were observed in most of cases, relatively to control group, but there is no clear trend in these modifications.
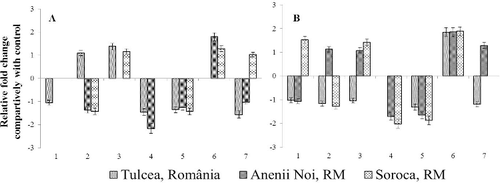
NPR1, the key regulator of SAR,[Citation10,Citation21] had minimal changes in transcriptional activity under the infection with broomrape comparatively with other studied genes. It was up-regulated only in case of MS-2067A and down-regulated in case of MS-2077A and MS-2091A. Obtained data showed that NPR1 does not modify expression in advanced stages of infection, being involved in early events which take place in SAR installation ().
Phenylalanine ammonia-lyase (PAL; E.C.4.3.1.5) is the key enzyme in the plant phenylpropanoid pathway, which catalyses the first step in the pathway (biotransformation of L-phenylalanine to trans-cinnamic acid and ammonia) and determines not only the accumulation of phytoalexins,[Citation22] but also ensures plant responses to biotic stresses, enhancing lignin synthesis for fortification of structural barriers.[Citation11,Citation23,Citation24] Similarly with NPR1, PAL gene demonstrated minor alterations in expression. Only in three cases PAL transcriptional activity was down-regulated under – four-fold (infected plants of MS-2091A genotype and non-symptomatic plants of MS-2039A) ().
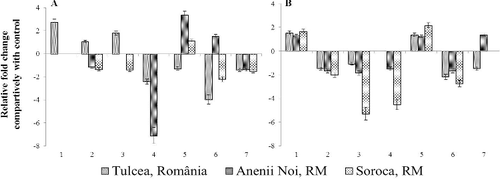
Plant defensin PDF1.2 is often considered as a classic marker for the induction of the JA-dependent defence-signalling pathway.[Citation21] Transcriptional activity of defensin was altered more than that of NPR1 and PAL genes. Thus, the expression fold change varies in limits from 8.67-fold (non-symptomatic plants of MS-2067 infected with Soroca population) up-regulation to −18.42-fold down-regulation (in plants of MS-2091A infected with Anenii Noi population) ().
PR5 proteins are large family of thaumatin-like proteins, which are present in both dicots and monocots and act at fungal membrane, increasing its permeability.[Citation25] PR5 proteins are considered as markers for SA-dependent SAR.[Citation2,Citation26]
Like defensin gene, PR5 mostly changed significantly its expression (e.g. PR5 was up-regulated up to 235-fold in non-symptomatic plants of MS-2039A infected with broomrape from Soroca). It has to be mentioned, that in common the highest values of transcript accumulations in responses to pathogen were detected for PR5 gene (3–235 folds), marker of SAR ().
According to the results obtained in this study, it was determined that in advanced stages of infection in defensive response participate PR5 and defensin genes, NPR1 and PAL probably being involved in early responses.
Summarizing the obtained results, it was found that at advanced stage of infection in cases of all three broomrape populations, around 30%–40% of cases have a transcriptional activity approximately equal with control group (within −1.5 – +1.5 limits), including 30% for the population from Soroca, 40% for the population from Anenii Noi and 46% for the population from Tulcea, which means that gene activity has returned to normal levels.
The most pronounced effect on expression of studied genes had population from Soroca. Comparatively, with two other populations, this population induced significant modification of transcriptional activity of studied genes in 70% of cases, including 27% in which it was up- or down-regulated more than four-fold ().
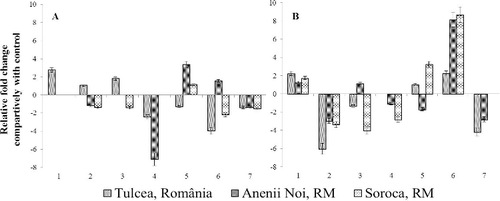
Figure 5. Number of differential expression cases for sunflower ROS-scavenging genes under the influence of broomrape infection.
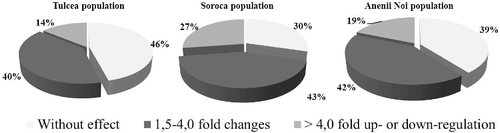
The most aggressive population of broomrape from Tulcea, Romania showed the least effect on gene expression. It was modified significantly only in 54% of cases. Thus, in 40% of cases expression was up- or down-regulated from 1.5 to 4.0 fold and in 14% more than four-fold.
Broomrape collected from Anenii Noi influenced expression of studied genes in 61% of cases. This population has intermediate effect comparatively with two other populations.
Obtained results showed that in the majority of cases gene transcriptional activity in advanced stages of broomrape infection was recovered or maintained at the normal level.
Conclusions
Based on data regarding attack frequency in controlled conditions and modification of gene expression in adaptation phase it was established that broomrape populations from Tulcea and Soroca regions are much more aggressive than population from Anenii Noi. Our studies revealed two genotypes MS-2161A and MS-2039A resistant to Anenii Noi population of broomrape. Moreover, MS-2161 was tolerant to Soroca population too.
Resistant genotypes (MS-2161A and MS-2039A) were characterized through higher stability of studied genes expression, being able to maintain and recover a normal level of metabolism in a wider range of stress factor values intensity.
Based on the fact that the expression of PR5 and defensin genes in infected plants was significantly different from the control, it has been suggested that these genes mostly participate in defensive response in advanced stages of infection, but NPR1 and PAL genes, which had lower alterations between control and infected plants, probably have been involved in early responses.
Studied genes showed different levels of transcript accumulation in function of the population of pathogen and investigated genotype. Performed investigation showed up-regulation of PAL and defensin genes or PR5 gene in sunflower resistant to broomrape, thus, suggesting implication of these genes in defence responses during infection and limitation of pathogen growth and development.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Anil K, Das SN, Podile AR. Induced defense in plants: a short overview. Proc Nat Acad Sci India B Biol Sci. 2014;84:669–679.
- Durrant W, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209.1
- Udovenko GV. Resistance of plants against abiotic stresses. Physiological bases of selection of plants. Theor Bases of Selection. 1995;II:293–352. Russian.
- López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593.
- Fofana B, Banks TW, McCallum B, et al. Temporal gene expression profiling of the wheat leaf rust pathosystem using cDNA microarray reveals differences in compatible and incompatible defence pathways. Int J Plant Genomics. 2007;2007:17542.
- Rioux R, Manmathan H, Singh P, et al. Comparative analysis of putative pathogenesis-related gene expression in two Rhizoctonia solani pathosystems. Curr Genet. 2011;57:391–408.
- Letousey P, De Zélicourt A, Vieira Dos Santos C, et al. Molecular analysis of resistance mechanisms to Orobanche cumana in sunflower. Plant Pathol. 2007;56:536–546.
- Radwan O, Mouzeyar S, Venisse JS, et al. Resistance of sunflower to the biotrophic oomycete Plasmopara halstedii is associated with a delayed hypersensitive response within the hypocotyls. J Exp Bot. 2005;56:2683–2693.
- Thomma BP, Penninckx IA, Broekaert WF, et al. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–68.
- Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552.
- Huang J, Gu M, Lai Z, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538.
- Datta K, Velazhahan R, Oliva N, et al. Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet. 1999;98:1138–1145.
- Chern M, Fitzgerald HA, Canlas PE, et al. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact. 2005;18:511–520.
- Schneiter AA, Miller J F. Description of sunflower growth stages. Crop Sci. 1981;21: 901–903.
- Kaya Y, Evci G, Pekcan V, et al. Determining new broomrape-infested areas, resistant lines and hybrids in Trakya region of Turkey. Helia. 2004;27:211–218.
- Untergasser A, Cutcutache I, Koressaar T, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408.
- Dospekhov B. Metodika polevogo opyta (s osnovami statisticheskoy obrabotki rezultatov issledovaniy) [The method of field experience (the basics of statistical processing of research results)]. Moscow: Agropromizdat Publ, 5th ed.;1985.
- Databank [ Internet]. Chisinau: National Bureau of Statistics of the Republic of Moldova [ cited 2015 Sept 28]. Available from: http://statbank.statistica.md/pxweb/Dialog/Saveshow.asp
- Echevarría-Zomeño S, Pérez-de-Luque A, Jorrín J, et al. Pre-haustorial resistance to broomrape (Orobanche cumana) in sunflower (Helianthus annuus): cytochemical studies. J Exp Bot. 2006;57:4189–4200.
- Grant M, Lamb C. Systemic immunity. Curr Opin Plant Biol. 2006;9:414–420.
- Castillo Ruiz RA, Herrera C, Ghislain M, et al. Organization of phenylalanine ammonia lyase (PAL), acidic PR-5 and osmotin-like (OSM) defence-response gene families in the potato genome. Mol Genet Genomics. 2005;274:168–179.
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097.
- Ferrer JL, Austin MB, Stewart C Jr, et al. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem. 2008;46:356–370.
- Anžlovar S, Dermastia M. The comparative analysis of osmotins and osmotin-like PR-5 proteins. Plant Biol. 2003;5:116–124.
- Molinari S, Fanelli E, Leonetti P. Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol Plant Pathol. 2014;15:255–264.