ABSTRACT
The purpose of this study was molecular characterization of the antibiotic resistance profiles of some Enterococcus isolates obtained from different hospitals in Taif governorate in KSA. Out of the 89 bacterial isolates obtained, 12 isolates of Enterococcus spp. were subjected to fingerprinting based on repetitive sequence-based polymerase chain reaction (Rep-PCR), and tested their resistance/susceptibility against some antibiotics which are commonly used in KSA. They were identified using the specific primers for different antibiotic resistance genes of Enterococcus spp. as Tuf, VanC-1, VanC-2-VanC-3 genes and sequencing fragments of 16S rDNA gene. The obtained results indicated that about 58.3% of Enterococcus isolates were Enterococcus faecium, 16.6% were Enterococcus durans and 25.1% were other Enterococcus species. Sixty-seven per cent of the isolates had multi-drug resistance patterns against gentamicin, vancomycin, erythromycin, amoxicillin, cefazolin and tetracycline. Data on the prevalence and types of antibiotic resistance in Enterococcus species may be used to describe baseline antibiotic susceptibility profiles associated with Enterococcus spp. that were isolated from the hospitals’ environment. Some discrepancies were detected among the identification methods used, and the most reliable were the Tuf, VanC-1, VanC-2-VanC-3 genes, and 16S rDNA nucleotide sequencing of 12 Enterococcus isolates were deposited in Gene Bank under the accession numbers from KT366721 to KT366732, respectively. Selected isolates exhibited susceptibility to almost all studied antibiotics, and some virulence factors were detected by PCR. Finally, these Enterococcus isolates were molecularly characterized by Rep-PCR into a diverse genetic background. The data collected may also help to elucidate the role of hospitals in the transmission of antibiotic-resistant strains to human populations.
Introduction
Enterococci are ubiquitous bacteria that have a predominant habitat in the digestive tract of humans and animals, but may be present in soil, water's surface and in plants.[Citation1,Citation2] Enterococci have emerged as important nosocomial pathogens over the past decade,[Citation3,Citation4] ranking only second to staphylococci as a leading cause of nosocomial infections, it associated as hospital infections in the Saudi Arabia,[Citation5] and the first report of multi-drug resistance Enterococcus from Saudi Arabia was in 1993 from King Faisal Specialist Hospital, Riyadh.[Citation6] They frequently possess several specific traits that enable them to survive in the hospital environment, colonize patients and cause infections such as bacteraemia, peritonitis, endocarditis and urinary tract, wound and device-related infections.[Citation2,Citation7]
There are two major pathogenic species of Enterococcus in humans, E. faecalis and E. faecium, with occasional infections being caused by E. durans, E. gallinarum, E. casseliflavus, E. avium, E. hirae, E. mundtii and E. raffinosus.[Citation8,Citation9] In particular, Enterococcus species have emerged as multi-resistant nosocomial pathogens in immune compromised and critically ill patients. Multi-resistant strains have acquired virulence genes resulting in hospital adapted clones.[Citation10,Citation11] Of human clinical importance, E. faecium strains had a much higher prevalence of resistance to ciprofloxacin, tetracycline and nitrofurantoin than E. faecalis. E. faecalis strains had a low prevalence of resistance to antibiotics used to treat E. faecalis infections of both clinical and of agricultural relevance, excluding its intrinsic resistance patterns.[Citation1,Citation2]
The taxonomy of the genus Enterococcus has been exposed to considerable changes in recent years as a consequence of a progressive increase in the number of novel species.[Citation12] 16S rRNA gene sequencing and repetitive sequence-based polymerase chain reaction (Rep-PCR) analysis are among the most common techniques currently used for Enterococcus species identification.[Citation10,Citation12–15] However, 16S rRNA gene sequences have limited discriminating power for several closely related enterococcal species, e.g. the E. faecium species group.[Citation12,Citation16] Tuf gene sequence analysis [Citation2] and multiplex VanC-1, VanC-2, VanC-3 PCR,[Citation2,Citation17] have been used for identification of several Enterococcus species. So far, these molecular techniques are not yet used for routine identification. The use of protein coding gene sequence data for the determination of genomic relatedness is emerging as an alternative to overcome these problems.[Citation12,Citation18] The main target of this study was molecular characterizing of multi-drug resistance Enterococcus species isolated from different hospitals in Taif governorate in KSA.
Material and methods
Sample collection and growth
About 89 clinical samples of urine and stool swabs were collected from hospitalized patients in the Taif hospitals, KSA in nine months. Clinical samples were taken in aseptic conditions and were transported immediately to the microbial genetics laboratory at Biotechnology and Genetic Engineering Unit, Scientific Research Center, Taif University, KSA.
Isolation and purification of clinical bacterial isolates
Sterile dry swabs were used for streaking of clinical samples onto sterile Petri dishes containing nutrient agar media (Biolife, USA). Inoculated streaked dishes were incubated at 28 °C for 48 h. Single colonies were picked up by sterile inoculation needles and then sloped into cultures of nutrient agar media.
Antibiotic sensitivity test
About eight types of antibiotics, 10 µg gentamicin (Gm), 15 µg erythromycin (Er), 5 µg vancomycin (Va), 10 µg penicillin G (P), 30 mg amoxicillin (Am), 30 µg cefazolin (Cz), 30 µg Nalidixic acid (Na) and 30 µg tetracycline (Te) were used for disc diffusion bioassay.[Citation19] Clinical bacterial isolates suspensions were spread by sterile glass rods on the surface of nutrient agar media. Then, antibiotic discs (Bioanalyse®) were placed onto the surface of the inoculated nutrient agar plates. The plates were then incubated at 28 °C for 48 h, and then inhibition zones were observed.
Genomic DNA extraction and identification by PCR
The cell pellets from all isolates were used to extract genomic DNA using (Jena Bioscience, Germany) extraction kit following the manufacturer's instructions. The identification of enterococci was detected by PCR using specific primers targeted ().[Citation2,Citation17]
Table 1. Primers used in this study for identification and detection of resistance genes by PCR-based method.
Rep-PCR technique
For repetitive sequence analysis, PCR conditions for Enterococcus isolates in the present investigations were standardized. Seven repetitive sequence primers were used to amplify genomic DNA of the 12 isolates according to [Citation12,Citation15] BOX A1(5'-CTA CGG CAA GGC GAC GCT GAC G-3'), REP1R-I (5'-III ICG ICG ICA TCI GGC-3') (forward), REP2-I (5'-ICG ICT TAT CIG GCC TAC-3') (reverse), (GTG)5 (5'-GTG GTG GTGGTG GTG-3'), REP2-II (5'-GAGAGAGAGAGAGAGAA-3'), REP12-II (5'-AGAGAGAGAGAGAGAGC-3'), REP18-II (5'-ACACACACACACACACG-3') and REP19-II (5'-AGAGAGAGAGAGAGAGTT-3'). Following the experiments for optimization of component concentrations, PCR amplifications of repetitive sequence primers were carried out in 25 μL volume containing 1 μL (about 20 ng) of genomic DNA, 12.5 μL of Go Taq® Green Master Mix (Promega, USA), 1 μL of primer (20 pmol) and deionized distilled water (up to a total volume of 25 μL). For DNA amplification, the C1000TM Thermo Cycler Bio-Rad, Germany, was programmed under the conditions involving denaturation at 94 °C for 5 min; 30 cycles of denaturation at 94 °C for 1 min; primer annealing at 52 °C for 45 s; primer extension at 72 °C for 2.5 min and final extension step at 72 °C for 10 min.
Detection of antibiotic genes
The detection of resistance genes was conducted by PCR in all isolates of enterococci. The presence of genes vanA, aac(6′)-Ie-aph(2″)-Ia, erm(B) and tet(L) was found for vancomycin, gentamicin, erythromycin and tetracycline, respectively ().[Citation2] PCR reactions were carried out in a total volume of 25 μL containing 1× PCR buffer, deoxyribonucleotide triphosphate (dNTPs), 1 unit of Taq DNA polymerases, 10 pmol of each primer and the DNA template. PCR amplifications were performed in C1000™ Thermo Cycler (Bio-Rad). PCR conditions for mecA genes were: 95 °C for 5 min, followed by 30 cycles of 30 s at 94 °C, 30 s of annealing at 56 °C and 1 min of extension at 72 °C, followed by 7 min as final extension at 72 °C. Amplification products (10 μL) were analysed on 1.5% agarose gels stained with ethidium bromide and visualized by UV illumination and were photographed by a Bio-Rad Gel Doc 2000 device.
16S rDNA sequencing of 12 Enterococcus isolates
A single DNA fragment of about 1200 bp representing the 16S rDNA gene was amplified in all isolates according to Hassan and Ismail.[Citation14] This fragment was purified from these isolates using QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA) and sequenced using an Applied Bioscience model 3130A DNA sequencer in Scientific Research Center, Biotechnology and Genetic Engineering Unit, Taif University, KSA. The sequence reads were edited and assembled using the DNASTAR software (Lasergene, Madison, WI, USA). BLAST searches were conducted using the NCBI server of http://www.ncbi.nlm.nih.gov/blast/Blast.cgi. The 16S rDNA sequences for Enterococcus isolates were deposited in Gene Bank under the accession numbers KT366721, KT366722, KT366723, KT366724, KT366725, KT366726, KT366727, KT366728, KT366729, KT366730, KT366731 and KT366732.
Data analysis
In order to determine the genetic relationship among studied bacteria, Rep-PCR data were scored for presence (1) or absence (0) of the bands using Gene Tools software from Syngene. A simple matching coefficient was estimated by means of the Jaccard's coefficient to construct a similarity matrix. Cluster analysis and dendrogram were produced on the basis of the unweighted average pair group method using the NTSYS-PC Statistical Package.[Citation20]
Results and discussion
Molecular identification of the Enterococcus isolates
A total of 89 bacterial isolates that were collected from different hospitals in Taif governorate in KSA were investigated for the presence of enterococci. Conventional cultivation showed that 14.6% of them were harbouring Enterococcus spp. Genus specific PCR with detection of Tuf gene was in accordance with these preliminary results (). According to the molecular identification using PCR species-specific primers for genes VanC-1, VanC-2-VanC-3 and Ddl-E for E. gallinarum, E. casseliflavus E. faecium, respectively. The obtained results indicated that about 58.3% of Enterococcus isolates were E. faecium, 16.6% were E. durans and 25.1% were other Enterococcus species ((a)). The same results were obtained using sequencing fragments of 16S rDNA gene for all Enterococcus isolates. Enterococci are considered as opportunistic pathogens which can cause a variety of infections in patients that have severe underlying diseases or that are immunocompromised. The major responsible for human enterococcal infections is E. faecalis followed by E. faecium and with a much lower incidence by other enterococcal species.[Citation21]
Figure 1. Amplification of Tuf gene producing in Enterococcus isolates by single PCR with size of about 112 bp. First lane is 50 bp molecular weight markers.
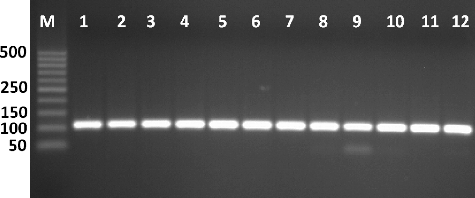
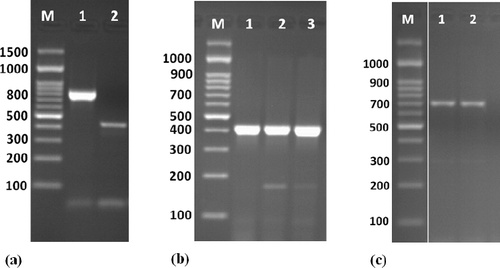
Figure 2. Amplification of some specific genes producing in Enterococcus isolates. (a) VanC-1 and VanC-2 genes specific for E. gallinarum and E. casseliflavus on isolates E8 and E9 by single PCR with size about of 822 and 439 bp, respectively. (b) Erm(B) gene specific for erythromycin resistance on isolates E6, E8 and E10 E9 by single PCR with size about of 405 bp. (c) Tet(L) gene specific for tetracycline resistance on isolates E8 and E10 E9 by single PCR with size about of 696 bp. First lane on each panel is 100 bp molecular weight markers.
The partial sequence of the 16S rRNA gene of the Enterococcus isolates is deposited in the GenBank database. Ribosomal operons are of great relevance for the study of bacterial evolution and phylogeny,[Citation14,Citation22] and sequencing of 16S rDNA has been widely used to reconstruct phylogenetic relationships of microorganisms [Citation14]. The partial 16S rDNA sequences from the Enterococcus isolates E1, E4, E5, E6, E7, E10 and E11 identified as E. faecium with accession numbers KT366721, KT366724, KT366725, KT366726, KT366727, KT366730 and KT366731, respectively. While, E2 and E3 identified as E. durans with accession numbers KT366722 and KT366723, respectively. Moreover, E8, E9 and E12 identified as E. gallinarum, E. casseliflavus and E. faecalis with accession numbers KT366728, KT366729 and KT366732, respectively. The 16S rRNA gene is very useful for discriminating the main groups of enterococci, i.e. E. avium, E. casseliflavus, E. cecorum, E. faecalis and E. faecium species groups.[Citation10,Citation12,Citation13] One example is the members of the E. faecium species group, i.e. E. faecium, E. hirae, E. durans, E. villorum, E. mundtii and E. ratti. The 16S rRNA genes of these species show similarities of 98.8%–99.7%.[Citation10,Citation12,Citation14]
Antibiotic susceptibility and virulence genes
A summary of the resistance among the Enterococcus isolates is reported in . The Enterococcus isolates displayed resistance to at least one antibiotic tested and were resistant to antibiotic agents, some of which were resistant to multiple drugs. The E. faecium isolates were mostly resistant to erythromycin (66.6%), followed by cefazolin (55.5%), tetracycline (50%), penicillin (42.8%) and completely sensitive against vancomycin. E. durans E2 was sensitive to all tested antibiotics, while E. durans E3 was mostly resistant to penicillin, amoxicillin, cefazolin and tetracycline. On the other hand, E. casseliflavus E9 was the only isolate resistant to gentamicin and Nalidixic acid. E. gallinarum E8 was resistant to erythromycin, penicillin, amoxicillin, cefazolin and tetracycline (). A total of 57.1% of E. faecium and 50% of E. durans isolates were resistant to multiple drugs. On the other hand, only three isolates, E6, E8 and E10, were resistance to erythromycin and harboured resistance genes erm(B) ((b)). The E. gallinarum E8 isolate was resistant to tetracycline harboured tet(L) gene. In spite of E. faecium E10 isolate harboured the tet(L) gene and presented tetracycline susceptibility phenotype. vanA gene detected only in E9 isolate that was resistant to vancomycin. Moreover, aac(6′)-Ie-aph(2′) was not detected in any Enterococcus isolates. Finally, some isolates harboured resistance genes to more than one antibiotic, the significant ones were tet(L)+/erm(B)+ to E. gallinarum E8 and E. faecium E10.
Table 2. Antimicrobial resistance profiles of 12 Enterococcus isolates.
Enterococci are intrinsically more resistant than other bacteria to antimicrobial agents commonly used in hospitals.[Citation4,Citation16] The danger of enterococcal infections becomes more serious in the light of increasing antimicrobial resistance, including resistance to multiple antibiotics and the possible transfer of resistance determinants to other bacterial genera.[Citation16] Some enterococcal species, particularly E. faecium, are inherently resistant to some penicillins, and in the past few years, they have also shown increased resistance to vancomycin, cephalosporins and aminoglycosides in nosocomial infections.[Citation4] Vancomycin and Synercid (quinupristin/dalfopristin) are often considered the last treatment available in serious, multi-drug resistant infections in humans.[Citation23,Citation24] Because of their role in human infections and their potential for harbouring antimicrobial resistance, it is important to identify genetic clones of enterococci. This has proven effective in clinical epidemiologic studies; however, genetic heterogeneity has been previously described for enterococci, particularly E. faecium, from hospital patients and environmental sources.[Citation16,Citation23]
Biodiversity of Enterococcus isolates by Rep-PCR analysis
Molecular markers are efficient tools for cultivar identification and estimation of relatedness through DNA fingerprinting. Rep-PCR markers have been used extensively in many different applications and in different microorganism species, because of the method’s simplicity.[Citation15,Citation25] Genomic diversity of Enterococcus was investigated by Rep-PCR analysis and was illustrated in and . The results indicated polymorphic numbers of the genetic bands, which were the electrophoretic products of PCR for Enterococcus isolates. Rep-PCR reactions were performed with 12 Enterococcus isolates collected from Taif hospitals, KSA. Eleven different Rep-PCR primers, which were preselected for their performance with Enterococcus DNA, were used. Out of the 11 primers, 7 retained for Rep-PCR analysis produced different fragment patterns with varied number of bands. The primers yielded a total of 120 distinct bands, 68.3.7% of them were considered as polymorphic and 31.6% of them were considered as monomorphic. displays the number of amplified fragments scored for each Rep-PCR primer. The amplified products were highly polymorphic among the 12 Enterococcus isolates.
Table 3. Polymorphism level detected by the seven repetitive sequence primers that have been used for Rep-PCR analysis.
Figure 3. Rep-PCR profile of 12 Enterococcus isolates generated with 6 repetitive sequence primers, a, b, c, e, d and f = REP1R-I, REP2-II, REP12-II, REP18-II, REP19-II and (GTG)5, respectively. First lane on each panel is 100 bp molecular weight markers.
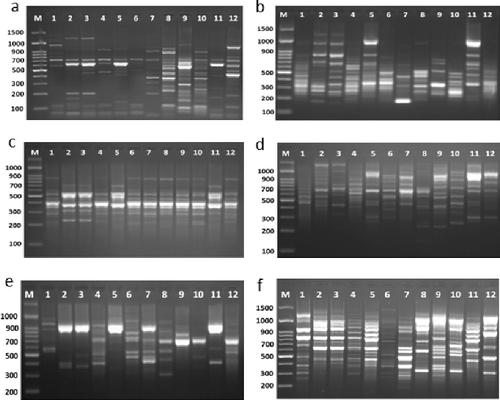
Obtained fragments from all analysis were enough for the identification and the evaluation of genetic similarities and designing the phylogenetic tree for these 12 Enterococcus isolates. The total number of bands, as shown in , varied from 13 different bands with primer REP2-II to 23 different bands with primer REP1R-I (). The total of monomorphic amplicons was 38 and the total of polymorphic amplicons was 82. It can be concluded from our study that Rep-PCR markers are effective in detecting similarity among Enterococcus isolates and they provide a potential tool for studying the inter-strain genetic similarity and the establishment of genetic relationships. In the case of REP2-II, primer has showed the lowest polymorphism and a total of seven fragments have shown polymorphism among the 12 Enterococcus isolates. The molecular size of the amplicon products ranged from 150 to 1650 bp. Also, this primer has recognized unique fragment in E2 and E3 Enterococcus isolates with about 1250 bp.
Figure 4. Dendrogram analysis among the 12 Enterococcus isolates based on the 7 repetitive sequence primers.
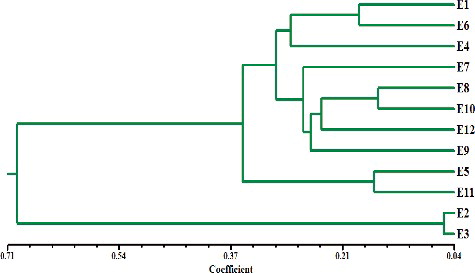
According to genetic similarity and intra-species differentiation, the 12 Enterococcus isolates were grouped into two different clusters with about 67% genetic similarity. Ten Enterococcus isolates were grouped in the first cluster and two Enterococcus isolates were grouped in the second cluster (). The overall genetic distance among Enterococcus isolates was relatively moderate. The smallest genetic distance (0.0927) was estimated between E1 isolates and E3 isolates. Among our Enterococcus isolates, E2 and E3 isolates relatively have high genetic distance (0.944), whereas, the genetic distances for E6 and E10 were marginally low. Rep-PCR was proved to be useful as genetic markers in Enterococcus fingerprinting.[Citation12,Citation13,Citation15,Citation18,Citation25,Citation26] Although major bands from Rep-PCR reactions are highly reproducible, minor bands can be difficult to repeat due to the nature of this PCR reaction and potential confounding effects associated with co-migration with other markers. In a previous study using different repetitive sequence primers, the discriminatory power of Rep-PCR and its ability to characterize strains were demonstrated. Rep-PCR primers also have been used in several previous studies and demonstrated to powerfully discriminate epidemiologically.[Citation15,Citation16,Citation26]
Conclusion
In conclusion, we demonstrated that the prevalence and types of antibiotic resistance of Enterococcus species may be used to describe baseline antibiotic susceptibility profiles associated with Enterococcus spp. The results indicate that all isolates harbour one or more of antibiotic resistance genes and that the PCR technique is a fast, practical and appropriate method for determining the presence of antibiotic resistance genes. The results may also help to elucidate the role of hospitals in the transmission of antibiotic-resistant strains to human populations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Giraffa G. Enterococci from foods. FEMS Microbiol Rev. 2002;26:163–171.
- Furlaneto-Maia L, Rocha K, Henrique F, et al. Antimicrobial resistance in Enterococcus sp isolated from soft cheese in Southern Brazil. Adv Microbiol. 2014;4:175–181.
- Vankerckhoven V, Van Autgaerden T, Vael C, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473–4479.
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278.
- Shorman M, Al-Tawfiq JA. Risk factors associated with vancomycin-resistant enterococcus in intensive care unit settings in Saudi Arabia. Interdisc Perspect Infect Dis. 2013;13:1–4.
- Qadri SMH, Qunibi WY, Al-Ballaa SR, et al. Vascomycin resistant Enterococcus: a case report and review of literature. Ann Saudi Med. 1993;13:289–293.
- Sava IG, Heikens E, Huebner J. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect. 2010;16:533–540.
- Ke D, Picard FJ, Martineau F, et al. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–3503.
- Frye JG, Jackson CR. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front Microbiol. 2013;4:135.
- Domig KJ, Mayer HK, Kneifel W. Methods used for the isolation, enumeration, characterisation and identification of Enterococcus spp. 2. Pheno- and genotypic criteria. Int J Food Microbiol. 2003;88:165–188.
- Garrido AM, Gálvez A, Pulido RP. Antimicrobial resistance in enterococci. J Infect Dis Ther. 2014;2:15–21.
- Pavel S, Vancanneyt M, Seman M, et al. Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol Lett. 2005;247:59–63.
- Wijetunge DS, Dunn P, Wallner E, et al. Fingerprinting of poultry isolates of Enterococcus cecorum using three molecular typing methods. J Vet Diagn Invest. 2012;24:1166–1171.
- Hassan MM, Ismail AI. Isolation and molecular characterization of some pathogenic mobile phone bacteria. Int J Biochem Biotechnol. 2014;3:516–522.
- Gaber A, Hassan MM, El-Dosoky ES, et al. In vitro antimicrobial comparison of Taif and Egyptian pomegranate peels and seeds extracts. J Appl Biol Biotech. 2015;3:012–017.
- Jackson CR, Spicer LM, Barrett JB, et al. Application of multiplex PCR, pulsed-field gel electrophoresis (PFGE), and BOX-PCR for molecular analysis of enterococci. In: Sameh Magdeldin, editor. Gel Electrophoresis - Principles and Basics; 2012. Available from: http://www.intechopen.com/books/gel-electrophoresis-principles-and-basics/application-ofmultiplex-pcr-pulsed-field-gel-electrophoresis-pfge-and-box-pcr-for-molecular-analysi
- Vakulenko SB, Donabedian SM, Vorkresenskiy AM, et al. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob Agents Chemother. 2003;47:1423–1426.
- Bedendo J, Pignatari AC. Typing of Enterococcus faecium by polymerase chain reaction and pulsed field gel electrophoresis. Braz J Med Biol Res. 2000;33:1269–1274.
- Franklin RC, Matthew AW, Jeff A, et al. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-second Informational Supplement. Wayne (PA): Clinical and Laboratory Standards Institute 2012. M100–S22.
- Rohlf FJ. NTSYS-PC numerical taxonomy and multivariate analysis system. Version 2.1. New York (NY): Exeter Software; 2000.
- Malek R, El-Attar A, Mohamed M, et al. Technological and safety properties display biodiversity among enterococci isolated from two Egyptian cheeses, Ras and Domiati. Int J Food Microbiol. 2012;153:314–322.
- Zarei M, Khajeh E, Shekarforoush S. Evaluation of the bacterial contamination of the Iranian currency notes. Iranian J Health Environ.2009;1:81–88.
- Boneca IG, Chiosis G. Vancomycin resistance: occurrence, mechanisms and strategies to combat it. Exp Opin Ther Targets. 2003;7:311–328.
- Hassan MM, Gaber A, Attia OA, et al. Molecular characterization of antibiotic resistance genes in pathogenic bacteria isolated from patients in Taif hospitals, KSA. Am J Phytomed Clin Ther. 2014;2:939–951.
- Bourdon N, Lemire A, Fines-Guyon M, et al. Comparison of four methods, including semi-automated rep-PCR, for the typing of vancomycin-resistant Enterococcus faecium. J Microbiol Methods. 2011;84:74–80.
- Weiss A, Domig KJ, Kneifel W, et al. Evaluation of PCR-based typing methods for the identification of probiotic Enterococcus faecium strains from animal feeds. Anim Feed Sci Technol. 2010;158:187–196.
