ABSTRACT
Ziziphus spina-christi (sidr) is a shrub, sometimes a tree, native to a vast area of Africa stretching from Mauritania to West Africa. In the Kingdom of Saudi Arabia, it is an exotic medicinal plant for many diseases. The aim of this study was to assess the genetic diversity within and among 34 accessions of Z. spina-christi collected from different regions of Saudi Arabia. The amplification of genomic DNA with 11 inter-simple sequence repeat (ISSR) primers yielded 105 scorable loci, of which 93.4% were found to be polymorphic. The observed number of alleles (na), effective number of alleles (ne), Nei's gene diversity (h) and genetic diversity estimated by Shannon's information index (I) were 1.93, 1.44, 0.26 and 0.41, respectively. The total genetic diversity, Ht (0.266 ± 0.0289) was close to the average intrapopulation genetic diversity, Hs (0.2199 ± 0.0216). A high level of gene flow (Nm = 2.37) between populations, reflecting high genetic differentiation (Gst = 0.1739). The analysis of molecular variance showed that the maximum value of genetic variation was found within populations (90%), whereas a low value of genetic variance was observed among populations. The analysis using the unweighted pair-group method with arithmetic averages clustered the population from Farasan Island as an out-group due to its geographical origin. The obtained results demonstrate that the ISSR markers may be used for evaluation of the genetic diversity due to their efficiency in revealing polymorphism even in closely related germplasm and may help in Ziziphus genome analysis.
Introduction
The genus Ziziphus belongs to the family Rhamnaceae with about 85 species. Ziziphus spina-christi (L.) Willd. (sidr) is one of the species of Zizyphus found in Saudi Arabia. It is a wild and cultivated plant distributed in the Middle East, Pakistan and in the North and East of Africa. Z. spina-christi can grow either as a tree or as a shrub. The leaves are short, the flowers are pedunculated and the yellow or red fruits are edible.[Citation1] Z. spina-christi is a medicinal plant and its leaf extract (peptide and cyclopeptide alkaloids) has neuroprotective and therapeutic roles against pentylenetetrazol convulsant effect.[Citation2] In Saudi Arabia, it is used for the treatment of many diseases like wounds, ulcers, etc. Mizrahi et al. [Citation3] reported that Ziziphus species are cultivated in hot and arid regions. Some pharmacological screening studies indicated that Z. spina-christi leaves appears to be a safe alternative to lower the blood glucose level.[Citation4] Regarding the reproductive biology of the Ziziphus genus, many studies have shown self-incompatibility and synchronous protandrous dichogamy.[Citation5,Citation6] Asatryan and Tel-Zur [Citation7] observed that three studied Ziziphus species, Z. jujuba, Z. mauritiana and Z. spina-christi, are xenogamous and self-incompatible. Therefore, an understanding of the genetic diversity of the Ziziphus genus is an important objective, since genetic diversity is a primordial component of biodiversity and it is related to the geographic distribution of the species.
In the assessment of genetic diversity, molecular markers based on DNA have many advantages. Among the polymerase chain reaction (PCR)-based marker techniques, inter-simple sequence repeats (ISSR) are one of the simplest and widely used markers.[Citation8] ISSR markers are considered as a dominant marker; two phenotypes can be distinguished at each locus following the Mendelian theory.[Citation9] Ratnaparkhe et al. [Citation10] suggested that ISSR markers could provide important information for genome analysis of plant species as a marker associated with disease resistance gene clusters. This marker involves amplification of genomic DNA by PCR using a primer typically attached at the 5’ or 3’ end with 1–4 arbitrary, often degenerate bases extended into the flanking sequences.[Citation11] ISSR markers offer great potential for differentiating closely related cultivars and the generated bands are very repeatable on duplicate samples.[Citation12] ISSR markers have been used for characterisation of germplasm,[Citation13] for assessment of genetic diversity and phylogenetic studies of different species,[Citation14,Citation15] for identification of DNA markers linked to agronomic traits [Citation10] and for plant breeding.[Citation16]
In this paper, we report the use of ISSR markers to assess the genetic diversity among and within wild populations of Z. spina-christi collected from different regions of Saudi Arabia.
Materials and methods
Sample collection
The leaf samples of 34 accessions of Z. spina-christi were collected from different regions of Saudi Arabia (). The young leaves were collected in silica gel for good quality and quantity of genomic DNA.
Table 1. Details of selected Z. spina-christi populations from Saudi Arabia.
DNA extraction
DNA was extracted from 0.2 g of leaf tissue using the modified CTAB (cetyl trimethylammonium) bromide method of Khan et al.[Citation17] The material was ground to a fine powder, using liquid nitrogen with a pestle and mortar. The fine powder of each sample was transferred in a 2-mL Eppendorf tube and suspended in 700 μL of extraction buffer with 3% CTAB (w/v), 100 mmol/L Tris-HCl, pH 8, 2 mol/L NaCl, 25 mmol/L ethylenediaminetetraacetic acid (EDTA), pH 8, 3% β-mercaptoethanol (v/v) and 3% polyvinylpyrrolidone (w/v). The suspension was mixed for 5 min and incubated in a water bath at 65 ºC for 30 min. The mixture was then removed from the water bath, cooled and an equal volume of chloroform:isoamyl alcohol (24:1) was added. The suspension was mixed for 10 min, followed by centrifugation at 10,000 rpm for 10 min at room temperature. The supernatant was collected, transferred to another 1.5 mL tube and precipitated with 2/3 volume ice-cold isopropanol for 1 h at −20 °C. The DNA was pelleted by centrifugation at 10,000 rpm for 10 min at 4 °C. The pellet was washed with 70% alcohol, dried at room temperature and then suspended in Tris-EDTA (TE) buffer (10 mmol/L Tris-HCl, 1 mmol/L EDTA, pH 8). The isolated DNA was assayed by Nanodrop 8000 spectrophotometer (Thermo Scientific) and then used for PCR amplification.
PCR amplification
Amplification of genomic DNA was performed with ISSR primers (). The PCR bead (GE Healthcare, UK) was used for the amplification of DNA. Each reaction was performed in 25 μL reaction volume containing 20 μL of deionized sterile water, 4 μL of DNA (50 ng) and 1 μL of 10 pmol/L of ISSR primer. After mixing the PCR components, the reaction was carried out in an Applied Biosystems Thermal Cycler (Singapore) using the following conditions: initial denaturation at 94 ºC for 4 min, followed by 40 cycles of denaturation for 1 min, annealing step at 45 ºC for 1 min, extension at 72 ºC for 1 min and final extension at 72 ºC for 7 min. The amplified fragments were electrophoresed in 1.5% agarose gel (Hoefer, Richmond CA, USA). The gel was documented using a Syngene BIO IMAGING system (Ingenius L, UK).
Table 2. ISSR primers used for PCR amplification.
Data analysis
The data from the ISSR marker analysis was scored for presence (1) and absence (0) of bands. Faint and unclear bands were not counted. The POPGEN 32 software was used to measure the following parameters: observed number of alleles (na), effective number of alleles (ne), Nei's gene diversity (h) [Citation18], number of polymorphic loci (PL), percentage of polymorphic loci (PPL) and Shannon's information index (I).[Citation19] To analyse the genetic diversity in subdivided populations, we determined the total genetic diversity (Ht), intrapopulation genetic diversity (Hs) and gene differentiation (Gst). The amount of gene flow between populations was calculated using population differentiation [(Nm = 0.5 (1−Gst)/Gst], according to McDermott and McDonald.[Citation20] A dendogram was constructed based on the genetic distance, UPGMA (unweighted pair-group method with arithmetic averages) modified from the NEIGHBOR procedure of PHYLIP software version 3.5. Analysis of molecular variance (AMOVA) was performed on three levels: within populations, among populations and among regions, using the GenAlex cross platform package version 6.1 based on 999 permutations.[Citation21]
Results and discussion
Assessment of genetic diversity is very important for the conservation of plant genetic resources in their natural habitat. Fifteen ISSR primers were used to produce DNA fingerprint profiles. Eleven primers amplified 105 loci across 34 individuals. The remaining primers were considered unsuitable due to poor amplification. Out of these 105 loci, 99 loci were polymorphic, reflecting rich allelic diversity in the sampled populations. The size of the amplified bands ranged between 250 and 3000 bp (). The obtained results from the genetic polymorphism analysis in different populations of Z. spina-christi, such as number of observed alleles (na), effective number of alleles (ne), gene diversity (h), Shannon's information index (I), number of polymorphic loci (PL) and percentage of polymorphic loci (PPL), are presented in . The results showed that within the four studied populations, the number of observed alleles (na) ranged from 1.45 in the population from Farasan Island to 1.773 in the population from the At-taif region. The maximum number of effective alleles (1.525) was found in Population 1 (from At-taif) and the lowest number (1.252) was detected in Population 3 (from Farasan Island). Based on the (h) and (I) values, Population 3 showed a slightly lower genetic diversity, whereas the results for the genetic diversity in Population 2 (from Riyadh) and Population 4 (from Jizan) were convergent. This highly detected polymorphism is in agreement with the findings of Singh et al. [Citation22], who studied the genetic diversity of Z. mauritiana using ISSR markers. A previous study conducted on the screening of the genetic relationships in Chinese Ziziphus, using sequence-related amplified polymorphism (SRAP) markers showed a high polymorphism (98.28%) in the selected individuals.[Citation23] Our analysis of the genetic polymorphism obtained with ISSR markers demonstrated that the highest percentage of polymorphic loci (77.36%), largest number of polymorphic loci (82) and the highest gene diversity (h) and Shannon's information index (0.2967 and 0.4356) were found in Population 1 from At-taif region. The lowest values were observed for Population 3 from Farasan Island. Therefore, the sampled individuals from At-taif region could be considered to possess a higher genetic variation as compared to the other populations, therefore some topographical features of At-taif region (high altitude) might be involved. A large amount of variability was observed in these sampled populations, exhibiting high intraspecific genetic diversity. Our results are in agreement with the findings of Wang et al. [Citation24], through the study conducted on the genetic diversity of castor bean (Ricinus communis L.) based on ISSR markers. This high genetic diversity within each population might also be explained by the predominant dichogamic reproduction of this species. This suggestion is in line with the report of Asatryan and Tel-Zur [Citation7], who revealed that three Ziziphus species, Z. jujuba, Z. mauritiana and Z. spina-christi, were characterised by synchronous protandrous dichogamous flower development.
Figure 1. ISSR marker profile generated by primer 4. M, Molecular marker (100 bp DNA ladder, Solis Biodyne, Estonia); Lanes T1–T9 (At-taif); Lanes R1–R12 (Riyadh); Lanes F1–F5 (Farasan Island); Lanes G1–G8 (Jizan).
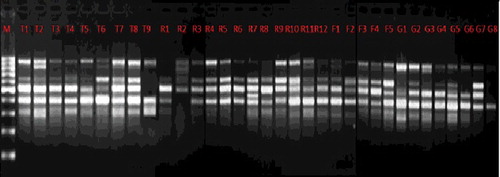
Table 3. Analysis of genetic polymorphism obtained with ISSR primers in different Z. spina-christi populations.
The analysis of the inter- and intra-population genetic structure () showed that the overall genetic diversity (Ht = 0.266 ± 0.0289) was similar to the intrapopulation genetic diversity (Hs = 0.2199 ± 0.0216). The genetic differentiation among the studied Z. spina-christi populations (Gst) was 0.1739 with effective gene flow observed between populations (Nm = 2.37). According to Nei [Citation25], Gst is classified as low when Gst < 0.05, medium when 0.05 < Gst < 0.15 and high when Gst > 0.15. Thus, the Gst coefficient of Z. spina-christi (Gst = 0.173) would be considered high. The Z. spina-christi species also had a gene flow Nm value greater than 1 (Nm = 2.37), indicating that our sampled populations were not subject to genetic drift.[Citation26] So the high genetic differentiation within populations may be caused by the outcrossing pollination phenomenon. Slatkin and Barton [Citation27] noted that, if the gene flow value is greater than 1, then it is reasonable to prevent substantial differentiation due to genetic drift. Compared to our results, low Nm and Gst have been detected in other Rhamnaceae species,[Citation28] in Liriodendron chinense (Magnoliaceae) and in Lilium cernuum (Liliaceae).[Citation29,Citation30]
Table 4. Nei's analysis of gene diversity in subdivided populations.
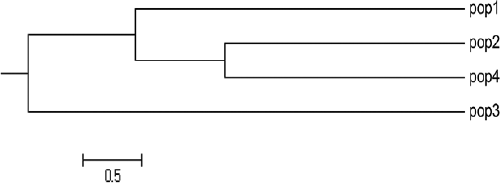
The matrix of the pairwise population PhiPT and Nei's genetic distance () also reveal the genetic differentiation between the 34 individuals from the different regions analysed in our study. This matrix showed that the highest PhiPT value and the genetic distance (0.138, 0.0908) were observed among Population 1 (At-taif) and Population 3 (Farasan Island), whereas the lowest values (0.072, 0.0405) were observed between Population 2 (Riyadh) and Population 4 (Jizan). This was also confirmed by the UPGMA results (), which showed separation of Population 3 individuals as an out-group consistent with their source (Island). Interestingly, Population 2 (Riyadh) and Population 4 (Jizan) were much closer despite the vast distance between them (127 km). These values of PhiPT and Nei's genetic distance were low when compared to other plants of different families, such as Rauvolfia serpentina, in which a comparatively larger genetic distance was reported. On the other hand, the individuals of this species were grouped into two clusters and each of them included populations close in geographical origin.[Citation31] When compared to another outcrossing species like Juniperus [Citation32], the pairwise PhiPT genetic distances of Z. spina-christi were the higher.
Table 5. Pairwise PhiPT values (above diagonal) and Nei's genetic distance (below diagonal) between selected groups.
Figure 2. UPGMA (based on Nei's genetic distance) dendogram showing the relationship between the four studied populations.
The AMOVA revealed that most of the genetic diversity occurred within populations (90%), while the genetic diversity among populations and among regions was 9% and 1%, respectively (). This indicates that Z. spina-christi is a relatively outcrossing species. The high value of within-population genetic variation in Z. spina-christi is in accordance with other studies in different plant species.[Citation33,Citation34] Overall, the obtained results demonstrate that ISSR markers are suitable for use in genome analysis and genetic diversity studies of Z. spina-christi, as they efficiently generated polymorphism even in closely-related germplasm.
Table 6. AMOVA of different selected populations of Z. spina-christi based on ISSR marker analysis.
Conclusions
The obtained results indicated that the Ziziphus genotypes investigated in this study have wide genetic diversity. High genetic differentiation was found mainly within populations, which may be caused by the outcrossing pollination phenomenon. Cluster analysis using the UPGMA method grouped the population from Farasan Island as an out-group, most probably due to its geographical origin. For better understanding of the genetic diversity of Z. spina-christi, future studies should focus on a larger number of populations and accessions collected from more geographical regions.
Acknowledgements
This study was supported by the Deanship of Scientific Research at King Saud University [project number RGP-014].
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chaudhary SA. Flora of the Kingdom of Saudi Arabia (Illustrated). Riyadh: National Agriculture and Water Research Center; 2001.
- Waggas AM., Al-Hasani RH. Neurophysiological study on possible protective and therapeutic effects of Sidr (Zizyphus spina-christi L.) leaf extract in male albino rats treated with pentylenetetrazol. SJBS. 2010;17:269–274.
- Mizrahi Y, Nerd A, Sitrit Y. New fruits for arid climates. In: Janick J, Whipkey A, editors. Trends in new crops and new uses. Alexandria (VA): ASHS Press; 2002. p. 378–384.
- Abdel-Zaher AO, Salim SY, Assaf MH, et al. Antidiabetic activity and toxicity of Zizyphus spina-christi leaves. J Ethnopharmacol. 2005;101:129–138.
- Weekley CW, Race T. The breeding system of Ziziphus celata Judd and DW Hall (Rhamnaceae), a rare endemic plant of the Lake Wales Ridge, Florida, USA: implications for recovery. Biol Conserv. 2001;100:207–213.
- Zietsman P, Botha F. Flowering of Ziziphus mucronata subsp. mucronata (Rhamnaceae): anthesis, pollination and protein synthesis. Bot Bull Acad Sin. 1992;33:33–42.
- Asatryan A, Tel-Zur N. Pollen tube growth and self-incompatibility in three Ziziphus species (Rhamnaceae). Flora. 2013;208:390–399.
- Vijayan K. Inter simple sequence repeat (ISSR) polymorphism and its application in mulberry genome analysis. Int J Indust Entomol. 2005;10:79–86.
- Tsumura Y, Ohba K, Strauss S. Diversity and inheritance of inter-simple sequence repeat polymorphisms in Douglas-fir (Pseudotsuga menziesii) and sugi (Cryptomeria japonica). Theor Appl Genet. 1996;92:40–45.
- Ratnaparkhe M, Tekeoglu M, Muehlbauer F. Inter-simple-sequence-repeat (ISSR) polymorphisms are useful for finding markers associated with disease resistance gene clusters. Theor Appl Genet. 1998;97:515–519.
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–183.
- Fang D, Roose M. Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor Appl Genet. 1997;95:408–417.
- Charters Y, Wilkinson M. The use of self-pollinated progenies as ‘in-groups’ for the genetic characterization of cocoa germplasm. Theor Appl Genet. 2000;100:160–166.
- Ajibade S, Weeden N, Chite S. Inter simple sequence repeat analysis of genetic relationships in the genus Vigna. Euphytica. 2000;111:47–55.
- Joshi S, Gupta V, Aggarwal R, et al. Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet. 2000;100:1311–1320.
- Reddy MP, Sarla N, Siddiq E. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica. 2002;128:9–17.
- Khan S, Qureshi MI, Kamaluddin TA, et al. Protocol for isolation of genomic DNA from dry and fresh roots of medicinal plants suitable for RAPD and restriction digestion. Afr J Biotechnol. 2007;6:175–178.
- Nei M. Molecular evolutionary genetics. New York (NY): Columbia University Press; 1987.
- Lewontin RC. The apportionment of human diversity. Evol Biol. 1995;6:381–398.
- McDermott JM, McDonald BA. Gene flow in plant pathosystems. Annu Rev Phytopathol. 1993;31:353–373.
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295.
- Singh A, Sharma P, Singh R, et al. Assessment of genetic diversity in Ziziphus mauritiana using inter-simple sequence repeat markers. J Plant Biochem Biotech. 2007;16:35–40.
- Li L, Peng J-y, Bai R-X. Analysis of the genetic relationships in Chinese Ziziphus with SRAP markers. Agri Sci China. 2010;9:1278–1284.
- Wang C, Li G-r, Zhang Z-y, et al. Genetic diversity of castor bean (Ricinus communis L.) in Northeast China revealed by ISSR markers. Biochem Syst Ecol. 2013;51:301–307.
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590.
- Karron JD. Patterns of genetic variation and breeding systems in rare plant species. In: Falk D, Holsinger K, editors. Genetics and conservation of rare plants. New York (NY): Oxford University Press; 1991. p. 87–98.
- Slatkin M, Barton NH. A comparison of three indirect methods for estimating average levels of gene flow. Evolution. 1989;43:1349–1368.
- Bacchetta G, Fenu G, Mattana E, et al. Genetic variability of the narrow endemic Rhamnus persicifolia Moris (Rhamnaceae) and its implications for conservation. Biochem Syst Ecol. 2011;39:477–484.
- Li K, Chen L, Feng Y, et al. High genetic diversity but limited gene flow among remnant and fragmented natural populations of Liriodendron chinense Sarg. Biochem Syst Ecol. 2014;54:230–236.
- Chung MY, Chung MG. Large effective population sizes and high levels of gene flow between subpopulations of Lilium cernuum (Liliaceae). Biochem Syst Ecol. 2014;54:354–361.
- Nair VD, Raj RPD, Panneerselvam R, et al. Assessment of diversity among populations of Rauvolfia serpentina Benth. Ex. Kurtz. from Southern Western Ghats of India, based on chemical profiling, horticultural traits and RAPD analysis. Fitoterapia. 2014;92:46–60.
- Teixeira H, Rodríguez-Echeverría S, Nabais C. Genetic diversity and differentiation of Juniperus thurifera in Spain and Morocco as determined by SSR. PloS One [Internet]. 2014 [cited 2016 Jan 12];9:e88996. Available from: http://dx.doi.org/10.1371/journal.pone.0088996.
- Pereira D, Corrêa RX, de Oliveira AC. Molecular genetic diversity and differentiation of populations of ‘somnus’ passion fruit trees (Passiflora setacea DC): Implications for conservation and pre-breeding. Biochem Syst Ecol. 2015;59:12–21.
- Jena SN, Verma S, Nair KN, et al. Genetic diversity and population structure of the mangrove lime (Merope angulata) in India revealed by AFLP and ISSR markers. Aquat Bot. 2015;120:260–267.
