ABSTRACT
The complete coding sequence of one tobacco (Nicotiana tabacum) gene, ascorbate oxidase (AO), was isolated by the application of reverse transcription-polymerase chain reaction. The tobacco AO gene consists of a 1722-bp open reading frame and encodes a protein of 573 amino acids. Sequence comparison analysis revealed that the tobacco AO protein shares high homology with the AO proteins of Lycopersicon esculentum (89%), Populus trichocarpa (75%), soybean (74%), castor bean (73%) and peach (73%). The prediction of transmembrane helices showed that tobacco AO might be a transmembrane protein. The expression profile was studied and the results indicated that the tobacco AO gene was diversely expressed in different tobacco tissues, including leaves, stem, roots and flowers. Our experiment laid the grounds for further research on this tobacco gene.
Keywords:
Introduction
Ascorbate oxidase (AO) is an apoplastic enzyme, which controls the redox state of the apoplastic ascorbate pool [Citation1–3]. AO is localized in the cell wall and catalyses the oxidation of ascorbate (AA) to the unstable radical monodehydroascorbate which quickly disproportionates to yield dehydroascorbate and AA [Citation4]. The lowered apoplast AA redox state in vivo, through increased AO expression in different plants, does not affect the expression levels of genes involved in AA recycling under normal growth conditions, but the plants show enhanced sensitivity to various oxidative stress-promoting agents [Citation5–7]. For example, transgenic tobacco plants over-expressing AO in their apoplast have altered ascorbate and glutathione redox states and increased sensitivity to ozone [Citation8]. Recent research also showed that AO RNAi tomato has increased stomatal conductance, leaf and fruit sugar content in the symplastic space, and increased fruit yield under unfavourable growth conditions [Citation9].
Although AO enzymes play important roles in plant biological processes, to date, the tobacco AO gene has not been reported yet. The aim of the present study was to isolate the complete mRNA sequence of the tobacco AO gene and subsequently perform tissue expression analysis for this gene. This will establish the primary foundation of understanding the AO tobacco gene.
Materials and methods
Samples collection, RNA extraction and first-strand cDNA synthesis
Tobacco plants (Chinese local variety Yunyan 85 obtained from Hongyun Honghe Tobacco (Group) Co., Ltd., Kunming, P.R. China) were grown in a naturally lit glasshouse with normal irrigation and fertilization. The tissues, including leaves, stem, root and flowers, were harvested and immediately frozen in liquid nitrogen and stored at −80 °C. Total RNA extraction and first-strand cDNA synthesis for these tissue samples were performed as described by Nian et al. [Citation10].
Isolation of the coding sequence
Reverse transcription-polymerase chain reaction (RT-PCR) was conducted to amplify the complete coding sequence of the tobacco AO gene, using the cDNA obtained from the pooled tissues above. The 20-µL RT-PCR reaction system was: 2.0 µL of cDNA, 2.0 µL of 2 mmol/L mixed deoxnucleoside triphosphates (dNTPs; Bioteke, Beijing, P.R. China), 2.0 µL of 10× Taq DNA polymerase buffer, 1.2 µL of 25 mmol/L MgCl2, 1.0 µL of 10 mmol/L forward primer, 1.0 µL of 10 mmol/L reverse primer, 2.0 units of Taq DNA polymerase (1 U/1 µL; Bioteke) and 9.8 µL of sterile water. The primers for tobacco AO gene isolation were designed based on the tobacco expressed sequence tag sequences (GeneBank numbers FG145178, FG153848 and AM823205), which are highly homologous with the coding sequence of the AO gene of Lycopersicon esculentum (). The PCR programme initially started with a 94 °C denaturation for 4 min, followed by 35 cycles of 94, 55 and 72 °C/1 min, then extension at 72 °C for 10 min and, finally, 4 °C to terminate the reaction (Thermalcycler Hema96TC, Zhuhai, P.R. China). Every PCR was repeated five times. The PCR products were analysed in an agarose gel (2.5%) containing ethidium bromide and then cloned into a PMD18-T vector and sequenced bidirectionally with the commercial fluorometric method (SHENGGONG, Shanghai, P.R. China). At least five independent clones were sequenced.
Table 1. PCR primers for tobacco AO gene isolation.
Quantitative real-time PCR (qRT-PCR) for tissue expression profile analysis
qRT-PCR was used to evaluate the level of mRNA of the AO gene on the ABI Prism 7300 Sequence Detection Systems (Applied Biosystems, Foster City, CA, USA). The 25-µL PCR reaction volume included 1 µL of SYBR™ Green real-time PCR Master Mix (Kangcheng Shengwu, Shanghai, P.R. China), 100 ng of cDNA template and 200 nmol/L of each primer. qRT-PCR procedures were: 95 °C for 3 min, 40 cycles of 95 °C for 15 s, optimal annealing temperature for each specific primer for 15 s () and 72 °C for 20 s. For each sample, the reactions were carried out in triplicate to ensure the reproducibility of the results. The AO gene expression levels were quantified relative to the expression of the reference gene actin (GenBank Accession number:. GQ339768) by employing the 2−ΔΔ Ct value model [Citation11].
Table 2. qRT-PCR primers for tobacco AO, actin genes and annealing temperature.
Sequence analysis
The gene prediction was performed by the GenScan software (http://genes.mit.edu/GENSCAN.html). The sequence comparison was conducted by BLAST (Basic Local Alignment Search Tool, http://www.ncbi.nlm.nih.gov/BLAST) and the ClustalW software (http://www.ebi.ac.uk/clustalw). The theoretical isoelectric point (pI) and molecular weight (Mw) of the protein were computed using the Compute pI/Mw Tool (http://www.expasy.org/tools/pi_tool.html). The transmembrane helices in the protein were predicted using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Results and discussion
Isolation of the tobacco AO gene
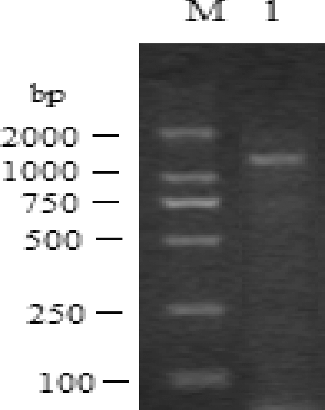
For the tobacco AO gene, the RT-PCR performed with pooled tissue cDNAs generated PCR products of 1722 bp ().
Sequence analysis
The cDNA BLAST analysis in the NCBI (National Center for Biotechnology Information) database revealed that this cDNA sequence was not homologous to any of the known tobacco genes and it was then deposited into the GenBank database (accession number: KF701478). The gene prediction that was carried out showed that this 1722-bp cDNA sequence is a single gene which encodes an AO protein of 573 amino acids (). The theoretical pI of the tobacco AO protein was 8.69 and its molecular weight was calculated to be 64,548.68.
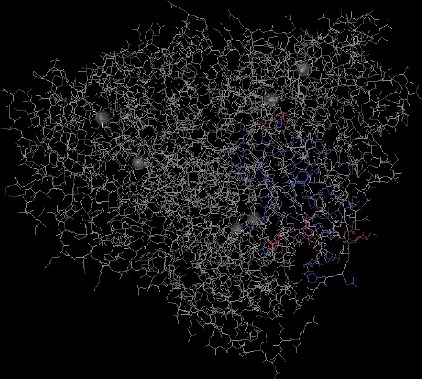
Further BLAST analysis of the tobacco AO protein revealed that the tobacco AO has high homology with the AO proteins of L. esculentum (accession number: NP_001234829, 89%), Populus trichocarpa (accession number: XP_002306323, 75%), soybean (accession number: XP_003524331, 74%), castor bean (accession number: XP_002528975, 73%) and peach (accession number: EMJ15865, 73%) (). Its conserved domain was identified as Cu-oxidase (). The three-dimensional structural evidence of the putative conserved domain is presented in
Figure 5. Three-dimensional structural evidence of the putative conserved domain of the tobacco AO protein.
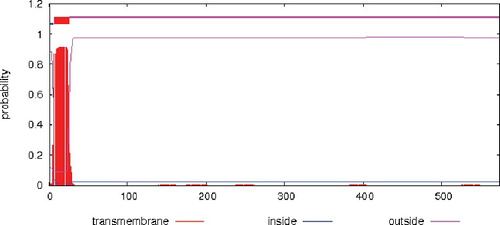
The prediction of transmembrane helices in the protein showed that the tobacco AO protein might be a transmembrane protein ().
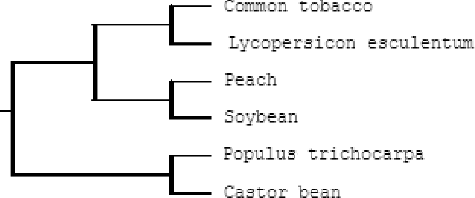
Based on the results from the multiple sequence alignment of AO proteins from different species, a phylogenetic tree was constructed as shown in . The phylogenetic tree indicated that the tobacco AO gene has a closer genetic relationship with that of L. esculentum.
Tissue expression profile
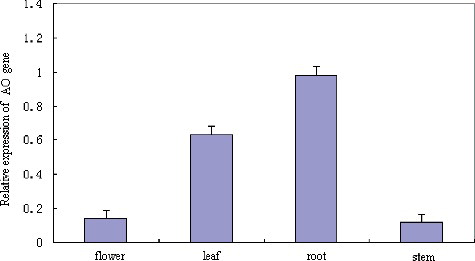
The results from the tissue expression profile analysis revealed that the tobacco AO gene was highly expressed in the root, moderately expressed in leaves and hardly expressed in flowers and stems ().
Comparative genomics research has revealed that there is extensive conservation in protein-coding regions in eukaryotes. Therefore, it has been speculated that such conservation might be expected in tobacco and L. esculentum, for they are both representatives of the Solanaceae family [Citation10,Citation12]. The results from the sequence comparison of AO genes indicated that the coding sequences of AO genes were indeed highly conserved in these two species. A similar observation has been made by Nian et al. [Citation10] regarding the isolation of another tobacco gene, TBG1 (β-galactosidase precursor) [Citation10]. This supports the suggestion that L. esculentum, whose genome database is already available [Citation13,Citation14], could serve as a model organism in studies aimed at isolation of other unknown tobacco genes based on the available coding sequence information of L. esculentum. Furthermore, the draft genomes of N. tabacum, recently announced, are expected to expand the use of N. tabacum as a model organism for functional genomics and biotechnology applications [Citation15].
Taken together, our results also demonstrate that the AO gene is diversely expressed in different parts of the tobacco plants. Since AO functions as an apoplastic enzyme which controls the redox state of the apoplastic ascorbate pool [Citation1–3], it could be concluded that the apoplastic enzyme activity of the protein encoded by the AO gene was diversely displayed in different tissues of the tobacco under the experimental conditions of this study.
Conclusions
To the best of our knowledge, this is the first study to report the isolation of a full-length tobacco AO cDNA. Sequence analysis revealed that the AO gene of tobacco has a closer genetic relationship with that of L. esculentum. The prediction of transmembrane helices showed that tobacco AO might be a transmembrane protein. The expression profile indicated that the tobacco AO gene was diversely expressed in different parts of the tobacco plants such as leaves, stems, roots and flowers. These results provide the basis for further research on this gene in tobacco.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Pignocchi C , Fletcher JM , Wilkinson JE , et al. The function of ascorbate oxidase in tobacco. Plant Physiol. 2003;132(3):1631–1641.
- Zhang Y , Han L , Ye Z , et al. Ascorbic acid accumulation is transcriptionally modulated in high-pigment-1 tomato fruit. Plant Mol Biol Rep. 2014;32:52–61.
- Höller S , Meyer A , Frei M . Zinc deficiency differentially affects redox homeostasis of rice genotypes contrasting in ascorbate level. J Plant Physiol. 2014;171:1748–1756.
- Fotopoulos V , Sanmartin M , Kanellis AK . Effect of ascorbate oxidase over-expression on ascorbate recycling gene expression in response to agents imposing oxidative stress. J Exp Bot. 2006;57:3933–3943.
- Chai L , Wang J , Fan Z , et al. Regulation of the flowering time of Arabidopsis thaliana by thylakoid ascorbate peroxidase. Afr J Biotechnol. 2014;11:7151–7157.
- Höller S , Ueda Y , Wu L , et al. Ascorbate biosynthesis and its involvement in stress tolerance and plant development in rice (Oryza sativ a L.). Plant Mol Biol. 2015;88:545–560.
- Chaudhary N , Agrawal SB . Cultivar specific variations in morphological and biochemical characteristics of mung bean due to foliar spray of ascorbic acid under elevated ozone. Acta Physiol Plant. 2014;36:1793–1803.
- Sanmartin M , Drogoudi PA , Lyons T , et al. Over-expression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta. 2003;216:918–928.
- Garchery C , Gest N , Do PT , et al. A diminution in ascorbate oxidase activity affects carbon allocation and improves yield in tomato under water deficit. Plant Cell Environ. 2013;36:159–175.
- Nian F , Zhang Y , Su X , et al. Isolation, sequence identification and tissue expression of a novel tobacco (Nicotiana tabacum) gene-TBG1. Int J Agric Sci Technol. 2013;1:57–61.
- Livak KJ , Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408.
- Hardison RC . Comparative genomics. PLoS Biol. 2003;1:156–160.
- Tomato Genome Consortium . The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641.
- PGSB Plant Genome and Systems Biology [Internet] . Neuherberg : Helmholtz Zentrum München Deutsches Forschungszentrum für Gesundheit und Umwelt; c2014. Tomato genome database [ cited 2016 Aug 24]. Available from: http://pgsb.helmholtz-muenchen.de/plant/tomato/index.jsp
- Sierro N , Battey JND , Ouadi S , et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. [Internet]. 2014 [ cited 2016 Aug 25];5:3833. Available from: http://www.nature.com/ncomms/2014/140508/ncomms4833/full/ncomms4833.html